Early diagnosis of SARS coronavirus infection by real time RT-PCR - PubMed (original) (raw)
Early diagnosis of SARS coronavirus infection by real time RT-PCR
Leo L M Poon et al. J Clin Virol. 2003 Dec.
Abstract
Background: A novel coronavirus was recently identified as the aetiological agent of Severe Acute Respiratory Syndrome (SARS). Molecular assays currently available for detection of SARS-coronavirus (SARS-CoV) have low sensitivity during the early stage of the illness.
Objective: To develop and evaluate a sensitive diagnostic test for SARS by optimizing the viral RNA extraction methods and by applying real-time quantitative RT-PCR technology.
Study design: 50 nasopharyngeal aspirate (NPA) samples collected from days 1-3 of disease onset from SARS patients in whom SARS CoV infections was subsequently serologically confirmed and 30 negative control samples were studied. Samples were tested by: (1) our first generation conventional RT-PCR assay with a routine RNA extraction method (Lancet 361 (2003) 1319), (2) our first generation conventional RT-PCR assay with a modified RNA extraction method, (3) a real-time quantitative RT-PCR assay with a modified RNA extraction method.
Results: Of 50 NPA specimens collected during the first 3 days of illness, 11 (22%) were positive in our first generation RT-PCR assay. With a modification in the RNA extraction protocol, 22 (44%) samples were positive in the conventional RT-PCR assay. By combining the modified RNA extraction method and real-time quantitative PCR technology, 40 (80%) of these samples were positive in the real-time RT-PCR assay. No positive signal was observed in the negative controls.
Conclusion: By optimizing RNA extraction methods and applying quantitative real time RT-PCR technologies, the sensitivity of tests for early diagnosis of SARS can be greatly enhanced.
Figures
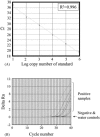
Fig. 1
Real-time quantitative RT-PCR assays for SARS-CoV. (A) Standard curve for quantitative analysis of ORF 1b of SARS-CoV. The threshold cycle (Ct) is the number of PCR cycles required for the fluorescent intensity of the reaction to reach a pre-defined threshold. The Ct is inversely proportional to the logarithm of the starting concentration of the input DNA. (B) An amplification plot of fluorescence intensity against the PCR cycle. The amplification curves of positive clinical samples, negative clinical samples and water controls are indicated. The X axis denotes the cycle number of a quantitative PCR assay. The Y axis denotes the fluorescence intensity over the background.
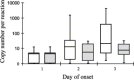
Fig. 2
Changes of viral loads of SARS-CoV in NPA samples collected at day 1-3 of disease onset. Open bar: viral loads of SARS-CoV from samples that were positive in the real-time quantitative PCR assay. Grey bar: Viral loads of SARS-Cov from samples that were positive in the real-time assay, but negative in the conventional RT-PCR assay. The upper and lower limits of the boxes and the lines across the boxes indicate the 75th and 25th percentiles and the median, respectively. The upper and lower horizontal bars indicate the 90th and 10th percentiles, respectively.
Similar articles
- A one step quantitative RT-PCR for detection of SARS coronavirus with an internal control for PCR inhibitors.
Poon LL, Wong BW, Chan KH, Leung CS, Yuen KY, Guan Y, Peiris JS. Poon LL, et al. J Clin Virol. 2004 Jul;30(3):214-7. doi: 10.1016/j.jcv.2003.12.007. J Clin Virol. 2004. PMID: 15135737 Free PMC article. - Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays.
Poon LL, Chan KH, Wong OK, Cheung TK, Ng I, Zheng B, Seto WH, Yuen KY, Guan Y, Peiris JS. Poon LL, et al. Clin Chem. 2004 Jan;50(1):67-72. doi: 10.1373/clinchem.2003.023663. Clin Chem. 2004. PMID: 14709637 Free PMC article. - Colonization of severe acute respiratory syndrome-associated coronavirus among health-care workers screened by nasopharyngeal swab.
Ho HT, Chang MS, Wei TY, Hsieh WS, Hung CC, Yang HM, Lu YT. Ho HT, et al. Chest. 2006 Jan;129(1):95-101. doi: 10.1378/chest.129.1.95. Chest. 2006. PMID: 16424418 Free PMC article. - [Detection of SARS coronavirus RNA].
Fukushi S. Fukushi S. Nihon Rinsho. 2005 Dec;63 Suppl 12:366-71. Nihon Rinsho. 2005. PMID: 16416820 Review. Japanese. No abstract available. - Molecular diagnosis of severe acute respiratory syndrome: the state of the art.
Mahony JB, Richardson S. Mahony JB, et al. J Mol Diagn. 2005 Nov;7(5):551-9. doi: 10.1016/S1525-1578(10)60587-9. J Mol Diagn. 2005. PMID: 16258152 Free PMC article. Review.
Cited by
- The Light and Shadow of Rapid Serological Tests for SARS-CoV-2 Infection: Results from a Study in a Large Emergency Department.
Loconsole D, Centrone F, Morcavallo C, Campanella S, Sallustio A, Quarto M, Procacci V, Chironna M. Loconsole D, et al. Int J Environ Res Public Health. 2020 Sep 7;17(18):6493. doi: 10.3390/ijerph17186493. Int J Environ Res Public Health. 2020. PMID: 32906596 Free PMC article. - Eco-Environmental Aspects of COVID-19 Pandemic and Potential Control Strategies.
Nazir R, Ali J, Rasul I, Widemann E, Shafiq S. Nazir R, et al. Int J Environ Res Public Health. 2021 Mar 27;18(7):3488. doi: 10.3390/ijerph18073488. Int J Environ Res Public Health. 2021. PMID: 33801704 Free PMC article. Review. - Challenges of SERS technology as a non-nucleic acid or -antigen detection method for SARS-CoV-2 virus and its variants.
Sitjar J, Liao JD, Lee H, Tsai HP, Wang JR, Liu PY. Sitjar J, et al. Biosens Bioelectron. 2021 Jun 1;181:113153. doi: 10.1016/j.bios.2021.113153. Epub 2021 Mar 9. Biosens Bioelectron. 2021. PMID: 33761416 Free PMC article. Review. - Confronting SARS: a view from Hong Kong.
Peiris JS, Guan Y. Peiris JS, et al. Philos Trans R Soc Lond B Biol Sci. 2004 Jul 29;359(1447):1075-9. doi: 10.1098/rstb.2004.1482. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15306392 Free PMC article. - The use of proteomics in the discovery of serum biomarkers from patients with severe acute respiratory syndrome.
Ren Y, He QY, Fan J, Jones B, Zhou Y, Xie Y, Cheung CY, Wu A, Chiu JF, Peiris JS, Tam PK. Ren Y, et al. Proteomics. 2004 Nov;4(11):3477-84. doi: 10.1002/pmic.200400897. Proteomics. 2004. PMID: 15378763 Free PMC article.
References
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Engl J Med. 2003;348:1967–1976. - PubMed
- Kaiser L., Briones M.S., Hayden F.G. Performance of virus isolation and Directigen Flu A to detect influenza A virus in experimental human infections. J Clin Virol. 1999;14:191–197. - PubMed
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S. A novel coronavirus associated with severe acute respiratory syndrome. New Engl J Med. 2003;348:1953–1966. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous