Inheritance of Polycomb-dependent chromosomal interactions in Drosophila - PubMed (original) (raw)
Inheritance of Polycomb-dependent chromosomal interactions in Drosophila
Frédéric Bantignies et al. Genes Dev. 2003.
Abstract
Maintenance of cell identity is a complex task that involves multiple layers of regulation, acting at all levels of chromatin packaging, from nucleosomes to folding of chromosomal domains in the cell nucleus. Polycomb-group (PcG) and trithorax-group (trxG) proteins maintain memory of chromatin states through binding at cis-regulatory elements named PcG response elements or cellular memory modules. Fab-7 is a well-defined cellular memory module involved in regulation of the homeotic gene Abdominal-B (Abd-B). In addition to its action in cis, we show here by three-dimensional FISH that the Fab-7 element leads to association of transgenes with each other or with the endogenous Fab-7, even when inserted in different chromosomes. These long-distance interactions enhance PcG-mediated silencing. They depend on PcG proteins, on DNA sequence homology, and on developmental progression. Once long-distance pairing is abolished by removal of the endogenous Fab-7, the derepressed chromatin state induced at the transgene locus can be transmitted through meiosis into a large fraction of the progeny, even after reintroduction of the endogenous Fab-7. Strikingly, meiotic inheritance of the derepressed state involves loss of pairing between endogenous and transgenic Fab-7. This suggests that transmission of nuclear architecture through cell division might contribute to inheritance of chromatin states in eukaryotes.
Figures
Figure 1.
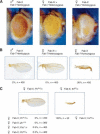
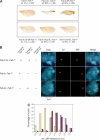
Pairing and PcG/trxG-dependent silencing mediated by the Fab-7 CMM in the Fab-X line. (A) Eye color phenotype of Fab-X flies grown at 25°C shows pairing-sensitive repression at the transgene. Genotypes are indicated above each panel. (B) Wing phenotype of Fab-X flies shows pairing-sensitive repression of the sd gene-flanking transgene insertion. The percentage of wings showing a sd phenotype at 29°C, and the number of wings analyzed are indicated. (C) Dependence of sd silencing on mutations in the PcG genes Pc, ph, Psc, and Pcl, and enhancement of silencing upon mutation of the trx gene.
Figure 2.
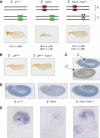
Silencing of sd in the Fab-X line depends on the presence of the endogenous Fab-7 element. (A) Schematic drawing illustrating the genotype of the lines used in the experiments. The two copies of chromosome X and chromosome 3 are shown. The endogenous Fab-7 at chromosome 3 is shown in green. The Fab-7 1 deletion is represented as an empty triangle. The transgenic copy inserted in chromosome X is shown in red. (B) sd phenotype in a loss-of function sd mutant in the presence or absence of endogenous Fab-7. (C) In situ hybridization of sd in embryos labeled with an anti-MSL-1 antibody to distinguish female (in blue) from male (in brown) embryos. (D) Repression of sd transcript levels in the Fab-X line (middle) compared with control w 1118 (left) and with Fab-X, Fab-7 1 (right) embryos. (E) Repression of sd transcript levels in third instar imaginal wing discs from the Fab-X line (middle) compared with control w 1118 (left) and with Fab-X, Fab-7 1 (right) larvae. sd is normally expressed in the wing pouch of the imaginal disc and it is repressed in Fab-X, but not in Fab-X, Fab-7 1 larvae.
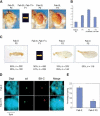
Figure 3.
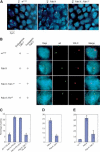
Long-range interactions between sd and the BX-C depend on sequence homology at Fab-7 and on the presence of PCL protein. (A) Two-color FISH in whole-mount female embryos. Examples of merged images of Dapi labeling (blue), the sd locus (green) and the BX-C (red) are shown. Projection of two _Z_-axis slices is shown. Embryo genotypes are indicated above each panel. Arrows indicate cases of colocalization between sd and the BX-C. (B) Single slices of individual nuclei show characteristic examples of data obtained in different lines. Dapi staining, the sd locus, the BX-C locus, and the merge of the three channels are shown. The genotypes and the presence of the endogenous or the transgenic Fab-7 copies are indicated. (C) Quantification of the percentage of colocalization of the two loci in stage 9-11 female embryos from different lines. The genotypes and the embryonic position of the analyzed nuclei are indicated below each bar. Error bars represent the standard deviation. At least 50 nuclei per embryo from four to sixembryos were analyzed in each experiment. (D) Same quantification as C, but from an independent experiment comparing female Fab-X embryos with mutant Fab-X; Pcl 10 embryos analyzed at stage 13-15 of development. (E) Quantification of long-distance Fab-7 pairing in third instar larval wing imaginal discs, measured as percentage of colocalization of the two loci in the different lines.
Figure 4.
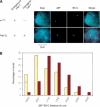
Long-range interactions between a Fab-7 transgene inserted at chromosome 2L and the BX-C. (A) Comparison of FISH signals from control w 1118 embryos and Fab-2L embryos at stage 9-11. (B) Quantitative analysis of the distribution of three-dimensional distances between the two loci in control w 1118 (brown bars) and Fab-2L (yellow bars) embryos. In the 0-0.5 μm distance range, two classes were distinguished, one class in which the two signals colocalized (11.0% for Fab-2L and 0.7% for w 1118, indicated by ticks within the bars), whereas in the other class, the two signals were close, but nonoverlapping.
Figure 5.
Association between transgenes carrying Fab-7. (A) In Fab-X; Fab-7 1 females, deletion of the endogenous Fab-7 derepresses sd (top, left). Strong repression is induced by introduction of a second Fab-7 transgene either in chromosome 2L (top, middle) or in chromosome 3R (top, right). Conversely, no repression is observed in the presence of the control pU/l5 transgene without CMM (bottom, left), or in the presence of a transgene carrying the bxd CMM (bottom, right). sd is slightly repressed upon introduction of a second transgene containing the Mcp CMM (bottom, middle). (B) Two-color FISH in female embryos carrying two Fab-7 transgenes, in the Fab-X,2L; Fab-7 1 line (top). Control female embryos are shown, carrying only one source of Fab-7, either at the sd locus (middle) or at the 38F locus in chromosome 2L (bottom). (C) Quantitative analysis of the distribution of three-dimensional distances between the two loci in control lines Fab-X; Fab-7 1 (blue bars) and Fab-2L; Fab-7 1 (brown bars) vs. the Fab-X,2L; Fab-7 1 line (yellow bars).
Figure 6.
Meiotic inheritance of chromatin states. (A) Inheritance of derepressed chromatin states in the Fab-2L line. Expression of the white gene is silenced in Fab-2L flies (F0). These flies were crossed with Fab-2L; Fab-7 1 in order to obtain a F1 progeny homozygous for the transgene and heterozygous for the endogenous Fab-7 copy. Females of this F1 generation were crossed with Fab-2L males. In the F2 generation, 50% of the progeny [identified by the absence of abdominal phenotypes (Gyurkovics et al. 1990) induced by the endogenous deletion of _Fab-7_] was genetically equivalent to the F0 (Fab-2L) generation, but was still derepressed. Derepression was maintained upon recrossing F2 Fab-2L flies (F3 generation). (B) Quantitation of eye pigment levels in control Fab-2L flies (F0 generation), in a F1 progeny homozygous for the transgene and heterozygous for the endogenous Fab-7 copy (Fab-2L; Fab-7 1/+), and in Fab-2L flies of the F2 generation obtained from a cross of Fab-2L; Fab-7 1/+ females with Fab-2L males (F2 via female) or from the reciprocal cross (F2 via male). (C) Inheritance of chromatin states in the Fab-X line. A similar crossing scheme was applied as in A and silencing of sd was analyzed. Genotypes, percentages of wings showing a sd phenotype, and numbers of wings analyzed are indicated. Two independent experiments are shown. sd derepression is inherited in F2 and F3 generation flies. (D) Two-color FISH in Fab-X F3 female embryos layed from F2 generation derepressed Fab-X flies (bottom), or from control Fab-X female embryos (top). (E) Quantification of the percentage of colocalization between the two loci in control Fab-X embryos (see also Fig. 3C for an independent experiment in the same condition), and Fab-X F3 embryos.
Figure 7.
A PcG body-hopping model for establishment of long-range chromatin-pairing interactions. (A) Nuclei are represented in a Rabl configuration (Rabl 1885), in which centromeres are assembled near the apical pole of the nucleus, pointing toward the surface of the embryo, whereas telomeres point toward the opposite pole. Two chromosomal arms projecting from centromeric heterochromatin toward the basal pole of the nucleus are shown. PcG target genes are postulated to locate at PcG bodies (in green). Each PcG body may contain one or several of these genes. Two loci containing homologous CMM are depicted in red. CMM located at PcG bodies may thus be constrained, but they may still be capable of exploring the nuclear territory in the neighborhood of their PcG body (dashed circle around the CMM). (B) Chromosome movements may lead to occasional PcG body contacts. During these contacts, CMM may move from a PcG body into another one (the old CMM locations are shown in orange). (C) This step may be repeated several times and may thus allow a CMM to explore significant areas in the cell nucleus, containing preferentially other PcG target genes. (D) This PcG body-hopping process may favor establishment and maintenance of specific interactions between homologous CMM.
Similar articles
- The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis.
Cavalli G, Paro R. Cavalli G, et al. Cell. 1998 May 15;93(4):505-18. doi: 10.1016/s0092-8674(00)81181-2. Cell. 1998. PMID: 9604927 - Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1.
Déjardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M, Locker D, Cavalli G. Déjardin J, et al. Nature. 2005 Mar 24;434(7032):533-8. doi: 10.1038/nature03386. Nature. 2005. PMID: 15791260 - A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development.
Maurange C, Paro R. Maurange C, et al. Genes Dev. 2002 Oct 15;16(20):2672-83. doi: 10.1101/gad.242702. Genes Dev. 2002. PMID: 12381666 Free PMC article. - Epigenetic inheritance of chromatin states mediated by Polycomb and trithorax group proteins in Drosophila.
Déjardin J, Cavalli G. Déjardin J, et al. Prog Mol Subcell Biol. 2005;38:31-63. doi: 10.1007/3-540-27310-7_2. Prog Mol Subcell Biol. 2005. PMID: 15881890 Review. - Polycombing the genome: PcG, trxG, and chromatin silencing.
Pirrotta V. Pirrotta V. Cell. 1998 May 1;93(3):333-6. doi: 10.1016/s0092-8674(00)81162-9. Cell. 1998. PMID: 9590168 Review. No abstract available.
Cited by
- Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs.
Chetverina D, Savitskaya E, Maksimenko O, Melnikova L, Zaytseva O, Parshikov A, Galkin AV, Georgiev P. Chetverina D, et al. Nucleic Acids Res. 2008 Feb;36(3):929-37. doi: 10.1093/nar/gkm992. Epub 2007 Dec 17. Nucleic Acids Res. 2008. PMID: 18086699 Free PMC article. - The roles of inducible chromatin and transcriptional memory in cellular defense system responses to redox-active pollutants.
Weinhouse C. Weinhouse C. Free Radic Biol Med. 2021 Jul;170:85-108. doi: 10.1016/j.freeradbiomed.2021.03.018. Epub 2021 Mar 28. Free Radic Biol Med. 2021. PMID: 33789123 Free PMC article. Review. - Multi-Scale Organization of the Drosophila melanogaster Genome.
Peterson SC, Samuelson KB, Hanlon SL. Peterson SC, et al. Genes (Basel). 2021 May 27;12(6):817. doi: 10.3390/genes12060817. Genes (Basel). 2021. PMID: 34071789 Free PMC article. Review. - Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila.
Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. Melnikova L, et al. Proc Natl Acad Sci U S A. 2004 Oct 12;101(41):14806-11. doi: 10.1073/pnas.0403959101. Epub 2004 Oct 1. Proc Natl Acad Sci U S A. 2004. PMID: 15465920 Free PMC article. - Chromatin changes in Anopheles gambiae induced by Plasmodium falciparum infection.
Ruiz JL, Yerbanga RS, Lefèvre T, Ouedraogo JB, Corces VG, Gómez-Díaz E. Ruiz JL, et al. Epigenetics Chromatin. 2019 Jan 7;12(1):5. doi: 10.1186/s13072-018-0250-9. Epigenetics Chromatin. 2019. PMID: 30616642 Free PMC article.
References
- Alkema M.J., Bronk, M., Verhoeven, E., Otte, A., van't Veer, L.J., Berns, A., and van Lohuizen, M. 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes & Dev. 11: 226-240. - PubMed
- Beisel C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857-862. - PubMed
- Breiling A., Turner, B.M., Bianchi, M.E., and Orlando, V. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412: 651-655. - PubMed
- Brown K.E., Guest, S.S., Smale, S.T., Hahm, K., Merkenschlager, M., and Fisher, A.G. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91: 845-854. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases