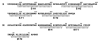Visualization of membrane protein domains by cryo-electron microscopy of dengue virus - PubMed (original) (raw)
. 2003 Nov;10(11):907-12.
doi: 10.1038/nsb990. Epub 2003 Oct 5.
Affiliations
- PMID: 14528291
- PMCID: PMC4148076
- DOI: 10.1038/nsb990
Visualization of membrane protein domains by cryo-electron microscopy of dengue virus
Wei Zhang et al. Nat Struct Biol. 2003 Nov.
Abstract
Improved technology for reconstructing cryo-electron microscopy (cryo-EM) images has now made it possible to determine secondary structural features of membrane proteins in enveloped viruses. The structure of mature dengue virus particles was determined to a resolution of 9.5 A by cryo-EM and image reconstruction techniques, establishing the secondary structural disposition of the 180 envelope (E) and 180 membrane (M) proteins in the lipid envelope. The alpha-helical 'stem' regions of the E molecules, as well as part of the N-terminal section of the M proteins, are buried in the outer leaflet of the viral membrane. The 'anchor' regions of E and the M proteins each form antiparallel E-E and M-M transmembrane alpha-helices, leaving their C termini on the exterior of the viral membrane, consistent with the predicted topology of the unprocessed polyprotein. This is one of only a few determinations of the disposition of transmembrane proteins in situ and shows that the nucleocapsid core and envelope proteins do not have a direct interaction in the mature virus.
Figures
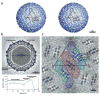
Figure 1
The dengue virus structure. (a) Stereo view of the viral surface at a resolution of 12.0 Å. The brown triangle demarcates the limits of one icosahedral asymmetric unit as defined by the five- and three-fold axes. Note the two protrusions per monomer corresponding to the glycosylation sites at Asn67 (yellow) and Asn153 (red). (b) A central cross section looking down an icosahedral three-fold axis, showing the polygonal shape of the membrane. The darkness of the shading is proportional to the magnitude of the cryo-EM density. Viral components are labeled. Maximum density heights are plotted below on a relative scale as a function of radius. (c) A radial cryo-EM density section at a radius of 185 Å, corresponding to the center of the lipid membrane, highlighting the herringbone arrangement of the three E dimers. The density is indicated in gray scale, with the highest density being the blackest. Shown also in brown is the limit of one icosahedral asymmetric unit. The boundaries of the E glycoprotein dimers are also indicated. The E dimer on the icosahedral twofold axis is red, whereas the monomers of the general-position dimer are blue and green. The transmembrane helices are viewed in cross section and marked for the green monomer according to the nomenclature of Figure 2.
Figure 2
Secondary structural predictions based on the primary sequences of the E and M stem-anchor regions. Helical coils represent the E stem (E-H1, E-H2) and transmembrane anchor (E-T1, E-T2) and the M stem (M-H) and transmembrane anchor (M-T1, M-T2) regions.
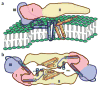
Figure 3
Stereoscopic diagrams showing the fit of the Cα backbones for the E and M regions into the cryo-EM density (gray) of the outer lipid (green) leaflet associated with the E dimer on the icosahedral two-fold axis. E ectodomains I, II and III are red, yellow and blue, respectively; stem-anchor region of E, cyan; M protein, orange; cryoEM density of the lipid bilayers, green. The stem and transmembrane helices are labeled with the nomenclature shown in Figure 2. Contour levels are chosen arbitrarily. The contour level for the lipid (green) is lower than that for the protein (gray). (a) Side view showing E and M monomers. (b) Enlarged view of a with a +50° rotation about the vertical axis to more clearly show the fit of E-H1 into the density. (c) Enlarged view of a with a –20° rotation about the vertical axis to more clearly show the fit of E-H2 and M-H into the density. (d) Top view of helices E-H1, E-H2 and M-H.
Figure 4
Diagrams of the dengue virus ectodomain and transmembrane domain proteins. The volume occupied by the ectodomain of an E monomer is pink (domain I), yellow (domain II) and lilac (domain III). The stem and anchor helices of E and M are blue and orange, respectively. Helices are identified by the nomenclature shown in Figure 2. CS represents the conserved sequence between E-H1 and E-H2. (a) View as in Figure 3a. (b) View as in Figure 3d with the superimposed E ectodomain homodimer.
Similar articles
- Structures of immature flavivirus particles.
Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Zhang Y, et al. EMBO J. 2003 Jun 2;22(11):2604-13. doi: 10.1093/emboj/cdg270. EMBO J. 2003. PMID: 12773377 Free PMC article. - Membrane curvature in flaviviruses.
Zhang W, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Zhang W, et al. J Struct Biol. 2013 Jul;183(1):86-94. doi: 10.1016/j.jsb.2013.04.005. Epub 2013 Apr 18. J Struct Biol. 2013. PMID: 23602814 Free PMC article. - Structure of West Nile virus.
Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Mukhopadhyay S, et al. Science. 2003 Oct 10;302(5643):248. doi: 10.1126/science.1089316. Science. 2003. PMID: 14551429 No abstract available. - Structures of viral membrane proteins by high-resolution cryoEM.
Zhou ZH. Zhou ZH. Curr Opin Virol. 2014 Apr;5:111-9. doi: 10.1016/j.coviro.2014.04.001. Epub 2014 May 3. Curr Opin Virol. 2014. PMID: 24799302 Free PMC article. Review. - Assembly and budding of influenza virus.
Nayak DP, Hui EK, Barman S. Nayak DP, et al. Virus Res. 2004 Dec;106(2):147-65. doi: 10.1016/j.virusres.2004.08.012. Virus Res. 2004. PMID: 15567494 Free PMC article. Review.
Cited by
- Dengue virus receptor.
Hidari KI, Suzuki T. Hidari KI, et al. Trop Med Health. 2011 Dec;39(4 Suppl):37-43. doi: 10.2149/tmh.2011-S03. Epub 2011 Aug 6. Trop Med Health. 2011. PMID: 22500135 Free PMC article. - Mutagenesis of the DI/DIII linker in dengue virus envelope protein impairs viral particle assembly.
de Wispelaere M, Yang PL. de Wispelaere M, et al. J Virol. 2012 Jul;86(13):7072-83. doi: 10.1128/JVI.00224-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532681 Free PMC article. - Inhibition of the Hantavirus Fusion Process by Predicted Domain III and Stem Peptides from Glycoprotein Gc.
Barriga GP, Villalón-Letelier F, Márquez CL, Bignon EA, Acuña R, Ross BH, Monasterio O, Mardones GA, Vidal SE, Tischler ND. Barriga GP, et al. PLoS Negl Trop Dis. 2016 Jul 14;10(7):e0004799. doi: 10.1371/journal.pntd.0004799. eCollection 2016 Jul. PLoS Negl Trop Dis. 2016. PMID: 27414047 Free PMC article. - Virus membrane-fusion proteins: more than one way to make a hairpin.
Kielian M, Rey FA. Kielian M, et al. Nat Rev Microbiol. 2006 Jan;4(1):67-76. doi: 10.1038/nrmicro1326. Nat Rev Microbiol. 2006. PMID: 16357862 Free PMC article. Review.
References
- Werten PJL, et al. Progress in the analysis of membrane protein structure and function. FEBS Lett. 2002;529:65–72. - PubMed
- Unger VM, Kumar NM, Gilula NB, Yeager M. Projection structure of a gap junction membrane channel at 7 Å resolution. Nat Struct Biol. 1997;4:39–43. - PubMed
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electroncrystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. - PubMed
- Cockburn JJB, Bamford JKH, Grimes JM, Bamford DH, Stuart DI. Crystallization of the membrane-containing bacteriophage PRD1 in quartz capillaries by vapour diffusion. Acta Crystallogr D. 2003;59:538–540. - PubMed
- Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, Pennsylvania, USA: 2001. pp. 991–1041.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources