The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses - PubMed (original) (raw)
The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses
Willemijn Vader et al. Proc Natl Acad Sci U S A. 2003.
Abstract
In patients with celiac disease, inflammatory T cell responses to HLA-DQ2-bound gluten peptides are thought to cause disease. Two types of HLA-DQ2 molecules exist, termed HLA-DQ2.5 and HLA-DQ2.2. Whereas HLA-DQ2.5 predisposes to celiac disease, HLA-DQ2.2 does not. We now provide evidence that the disease-associated HLA-DQ2.5 molecule presents a large repertoire of gluten peptides, whereas the non-disease-associated HLA-DQ2.2 molecule can present only a subset of these. Moreover, gluten presentation by HLA-DQ2 homozygous antigen-presenting cells was superior to presentation by HLA-DQ2/non-DQ2 heterozygous antigen-presenting cells in terms of T cell proliferation and cytokine secretion. Gluten presentation by HLA-DQ2.5/2.2 heterozygous antigen-presenting cells induced intermediate T cell stimulation. These results correlated with peptide binding to the antigen-presenting cells. Finally, we demonstrate that HLA-DQ trans dimers formed in HLA-DQ2.5/2.2 heterozygous individuals have properties identical with HLA-DQ2.5 dimers. Our findings explain the strongly increased risk of disease development for HLA-DQ2.5 homozygous and HLA-DQ2.2/2.5 heterozygous individuals, and they are indicative of a quantitative model for disease development, where HLA-DQ expression and the available number of T cell-stimulatory gluten peptides are critical limiting factors. This model may have important implications for disease prevention.
Figures
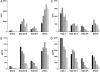
Fig. 1.
Gene dose effect in stimulation of gluten-specific T cell clones. Stimulation of two gluten-specific T cell clones with the gluten epitopes glia-α9(A and B) and glt-17 (C and D) is shown. Five concentrations of the epitopes were tested in the range from 0.2 μg/ml (light-gray bars) to 20 μg/ml (black bars). Peripheral blood mononuclear cells (PBMC) of various HLA-DQ2+ healthy individuals were used as APC (x axis). The epitopes were tested as deamidated peptides. T cell stimulation was determined by measurement of proliferation (A and C) and IFN-γ production (B and D). Production of tumor necrosis factor α, IL-10, IL-5, and IL-4 correlated with the levels of IFN-γ secreted (data not shown). The percentage of HLA class II-expressing cells in the PBMCs was determined by FACS analysis and was similar for each donor (data not shown). These results are representative of three independent experiments using PBMCs from various donors.
Fig. 2.
Peptide binding to a panel of HLA-DQ combinations. Shown is the binding of a concentration range of the indicator peptides MHCIα46-63 (A and B) and glia-α9(C and D) to HLA-DQ molecules isolated from HLA-homozygous and -heterozygous EBV-transformed B cell lines. The results shown are representative of at least three independent experiments.
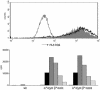
Fig. 3.
Functional expression of HLA-DQ2.5 in cis and in trans. (Upper) Absence of HLA-DQ2 on an HLA class II negative B cell line (thin line), and presence of HLA-DQ2 on the cell line transduced with HLA-DQα*0501 and DQβ*0201 (solid gray), and HLA-DQα*0501 and DQβ*0202 (black line). (Lower) T cell stimulation by the glia-α9 epitope in the context of the untransduced HLA class II-negative cell line (wt), and transduced cell lines with the HLA-DQ2 molecules in cis (DQα*0501 and DQβ*0201) and in trans (DQα*0501 and DQβ*0202). Five concentrations of the glia-α9 epitope were tested in the range from 0.2 μg/ml (light-gray bars) to 20 μg/ml (black bars).
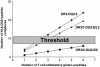
Fig. 4.
Model for the impact of the HLA-gene dose on the formation of immunogenic HLA-DQ2-gluten peptide complexes. With an increase in the number of available gluten peptides (x axis), the number of immunogenic HLA-DQ-peptide complexes (y axis) rises the strongest in HLA-DR3-DQ2.5-homozygous individuals, breaking through a hypothetical threshold when relatively few gluten peptides are present. HLA-heterozygous individuals need to recognize a larger group of gluten peptides before they reach the threshold. HLA-DQ2.5/2.2 individuals are in between.
Similar articles
- Design, synthesis and evaluation of high-affinity binders for the celiac disease associated HLA-DQ2 molecule.
Kapoerchan VV, Wiesner M, Hillaert U, Drijfhout JW, Overhand M, Alard P, van der Marel GA, Overkleeft HS, Koning F. Kapoerchan VV, et al. Mol Immunol. 2010 Feb;47(5):1091-7. doi: 10.1016/j.molimm.2009.10.036. Epub 2009 Dec 3. Mol Immunol. 2010. PMID: 19962195 - T-cell response to gluten in patients with HLA-DQ2.2 reveals requirement of peptide-MHC stability in celiac disease.
Bodd M, Kim CY, Lundin KE, Sollid LM. Bodd M, et al. Gastroenterology. 2012 Mar;142(3):552-61. doi: 10.1053/j.gastro.2011.11.021. Epub 2011 Nov 19. Gastroenterology. 2012. PMID: 22108197 - HLA-DQ2.5 genes associated with celiac disease risk are preferentially expressed with respect to non-predisposing HLA genes: Implication for anti-gluten T cell response.
Pisapia L, Camarca A, Picascia S, Bassi V, Barba P, Del Pozzo G, Gianfrani C. Pisapia L, et al. J Autoimmun. 2016 Jun;70:63-72. doi: 10.1016/j.jaut.2016.03.016. Epub 2016 Apr 12. J Autoimmun. 2016. PMID: 27083396 - Celiac disease--sandwiched between innate and adaptive immunity.
Stepniak D, Koning F. Stepniak D, et al. Hum Immunol. 2006 Jun;67(6):460-8. doi: 10.1016/j.humimm.2006.03.011. Epub 2006 Mar 30. Hum Immunol. 2006. PMID: 16728270 Review. - The molecular basis of celiac disease.
Koning F. Koning F. J Mol Recognit. 2003 Sep-Oct;16(5):333-6. doi: 10.1002/jmr.641. J Mol Recognit. 2003. PMID: 14523946 Review.
Cited by
- Apoptosis in response to microbial infection induces autoreactive TH17 cells.
Campisi L, Barbet G, Ding Y, Esplugues E, Flavell RA, Blander JM. Campisi L, et al. Nat Immunol. 2016 Sep;17(9):1084-92. doi: 10.1038/ni.3512. Epub 2016 Jul 25. Nat Immunol. 2016. PMID: 27455420 Free PMC article. - Coeliac disease and rheumatoid arthritis: similar mechanisms, different antigens.
Koning F, Thomas R, Rossjohn J, Toes RE. Koning F, et al. Nat Rev Rheumatol. 2015 Aug;11(8):450-61. doi: 10.1038/nrrheum.2015.59. Epub 2015 May 19. Nat Rev Rheumatol. 2015. PMID: 25986717 Review. - The landscape of host genetic factors involved in immune response to common viral infections.
Kachuri L, Francis SS, Morrison M, Wendt GA, Bossé Y, Cavazos TB, Rashkin SR, Ziv E, Witte JS. Kachuri L, et al. medRxiv [Preprint]. 2020 Sep 5:2020.05.01.20088054. doi: 10.1101/2020.05.01.20088054. medRxiv. 2020. PMID: 32511533 Free PMC article. Updated. Preprint. - HLA genotyping in pediatric celiac disease patients.
Stanković B, Radlović N, Leković Z, Ristić D, Radlović V, Nikčević G, Kotur N, Vučićević K, Kostić T, Pavlović S, Zukic B. Stanković B, et al. Bosn J Basic Med Sci. 2014 Aug 16;14(3):171-6. doi: 10.17305/bjbms.2014.3.28. Bosn J Basic Med Sci. 2014. PMID: 25172978 Free PMC article. - Production and molecular characterization of bread wheat lines with reduced amount of α-type gliadins.
Camerlengo F, Sestili F, Silvestri M, Colaprico G, Margiotta B, Ruggeri R, Lupi R, Masci S, Lafiandra D. Camerlengo F, et al. BMC Plant Biol. 2017 Dec 19;17(1):248. doi: 10.1186/s12870-017-1211-3. BMC Plant Biol. 2017. PMID: 29258439 Free PMC article.
References
- van de Wal, Y., Kooy, Y. M. C., Drijfhout, J. W., Amons, R. & Koning, F. (1996) Immunogenetics 44, 246-253. - PubMed
- Vartdal, F., Johansen, B. H., Friede, T., Thorpe, C. J., Stevanovic, S., Eriksen, J. E., Sletten, K., Thorsby, E., Rammensee, H. G. & Sollid, L. M. (1996) Eur. J. Immunol. 26, 2764-2772. - PubMed
- Molberg, O., McAdam, S. N., Korner, R., Quarsten, H., Kristiansen, C., Madsen, L., Fugger, L., Scott, H., Noren, O., Roepstorff, P., et al. (1998) Nat. Med. 4, 713-717. - PubMed
- van de Wal, Y., Kooy, Y., van Veelen, P., Pena, S., Mearin, L., Papadopoulos, G. & Koning, F. (1998) J. Immunol. 161, 1585-1588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials