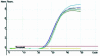A novel real-time quantitative PCR method using attached universal template probe - PubMed (original) (raw)
A novel real-time quantitative PCR method using attached universal template probe
Yuanli Zhang et al. Nucleic Acids Res. 2003.
Abstract
A novel real-time quantitative polymerase chain reaction (PCR) method using an attached universal template (UT) probe is described. The UT is an approximately 20 base attachment to the 5' end of a PCR primer, and it can hybridize with a complementary TaqMan probe. One of the advantages of this method is that different target DNA sequences can be detected employing the same UT probe, which substantially reduces the cost of real-time PCR set-up. In addition, this method could be used for simultaneous detection using a 6-carboxy-fluorescein-labeled UT probe for the target gene and a 5-hexachloro-fluorescein-labeled UT probe for the reference gene in a multiplex reaction. Moreover, the requirement of target DNA length for UT-PCR analysis is relatively flexible, and it could be as short as 56 bp in this report, suggesting the possibility of detecting target DNA from partially degraded samples. The UT-PCR system with degenerate primers could also be designed to screen homologous genes. Taken together, our results suggest that the UT-PCR technique is efficient, reliable, inexpensive and less labor-intensive for quantitative PCR analysis.
Figures
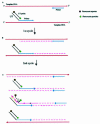
Figure 1
Schematic drawings of signal generation of the UT–PCR amplification. (A) The UT–PCR primer is composed of the 5′ end-attached UT sequence hybridized with the UT probe, and the 3′ end specifically hybridizes to the target sequence. (B) During the first cycle of the PCR amplification, the 3′ end of the UT–PCR primer is extended, generating a chimeric DNA fragment with the UT sequence on the 5′ end and newly synthetic target DNA on the 3′ end. (C) During the second cycle of the PCR amplification, the 3′ end of a free UT–PCR primer and the other primer anneal to available chimeric target DNA. The UT probe specifically anneals to the UT in the chimeric DNA fragments. Then, the 5′ exonuclease activity of DNA polymerase begins to hydrolyze the hybridized UT probe, and sets the reporter moiety free, thus generating a fluorescent signal. This amplification generates more chimeric DNA fragments.
Figure 2
Comparison of the amplification efficiency between the target DNA-specific primers and the same primer pairs with one attached with UT using SYBR Green I fluorogenic dye (Lectin gene, four replicates per reaction).
Figure 3
Sensitivity, precision and dynamic range of fluorogenic real-time PCR. Serial dilutions (10-fold) of transgenic maize Event 176 ranging from 0.01 to 100 ng were detected using a FAM-labeled fluorogenic primer (primer set 2, Table 2). (A) Amplification plot. (B) Initial DNA concentration versus Ct standard curve (_R_2 = 0.994, reaction efficiency = 0.99, three replicates per dilution).
Figure 3
Sensitivity, precision and dynamic range of fluorogenic real-time PCR. Serial dilutions (10-fold) of transgenic maize Event 176 ranging from 0.01 to 100 ng were detected using a FAM-labeled fluorogenic primer (primer set 2, Table 2). (A) Amplification plot. (B) Initial DNA concentration versus Ct standard curve (_R_2 = 0.994, reaction efficiency = 0.99, three replicates per dilution).
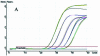
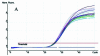
Figure 4
Multiplex fluorogenic PCR to detect the Invertase 1 and CryIA(b) gene using serial dilutions (10-fold) of transgenic maize Event 176. (A) Amplification plot of endogenous Invertase 1 gene. Each dilution contains 100 ng of total maize DNA. (B) Amplification plot of the transgenic CryIA(b) gene, serial dilutions of transgenic maize Event 176 ranging from 0.01 to 100 ng. (C) Initial Event 176 DNA concentration versus Ct standard curve. (D) Standard curve, plotting log (GMO amount) versus ΔCt (_R_2 = 0.993, reaction efficiency = 0.98, three replicates per dilution).
Figure 4
Multiplex fluorogenic PCR to detect the Invertase 1 and CryIA(b) gene using serial dilutions (10-fold) of transgenic maize Event 176. (A) Amplification plot of endogenous Invertase 1 gene. Each dilution contains 100 ng of total maize DNA. (B) Amplification plot of the transgenic CryIA(b) gene, serial dilutions of transgenic maize Event 176 ranging from 0.01 to 100 ng. (C) Initial Event 176 DNA concentration versus Ct standard curve. (D) Standard curve, plotting log (GMO amount) versus ΔCt (_R_2 = 0.993, reaction efficiency = 0.98, three replicates per dilution).
Figure 4
Multiplex fluorogenic PCR to detect the Invertase 1 and CryIA(b) gene using serial dilutions (10-fold) of transgenic maize Event 176. (A) Amplification plot of endogenous Invertase 1 gene. Each dilution contains 100 ng of total maize DNA. (B) Amplification plot of the transgenic CryIA(b) gene, serial dilutions of transgenic maize Event 176 ranging from 0.01 to 100 ng. (C) Initial Event 176 DNA concentration versus Ct standard curve. (D) Standard curve, plotting log (GMO amount) versus ΔCt (_R_2 = 0.993, reaction efficiency = 0.98, three replicates per dilution).
Figure 4
Multiplex fluorogenic PCR to detect the Invertase 1 and CryIA(b) gene using serial dilutions (10-fold) of transgenic maize Event 176. (A) Amplification plot of endogenous Invertase 1 gene. Each dilution contains 100 ng of total maize DNA. (B) Amplification plot of the transgenic CryIA(b) gene, serial dilutions of transgenic maize Event 176 ranging from 0.01 to 100 ng. (C) Initial Event 176 DNA concentration versus Ct standard curve. (D) Standard curve, plotting log (GMO amount) versus ΔCt (_R_2 = 0.993, reaction efficiency = 0.98, three replicates per dilution).
Figure 5
Screening of the CryIA(b) fragment in three lines of insect- resistant maizes (Bt11, Event 176, MON810) using degenerate UT–PCR primers (three replicates per sample).
Similar articles
- Event specific qualitative and quantitative polymerase chain reaction detection of genetically modified MON863 maize based on the 5'-transgene integration sequence.
Yang L, Xu S, Pan A, Yin C, Zhang K, Wang Z, Zhou Z, Zhang D. Yang L, et al. J Agric Food Chem. 2005 Nov 30;53(24):9312-8. doi: 10.1021/jf051782o. J Agric Food Chem. 2005. PMID: 16302741 - Detection and quantification of transgenes in grains by multiplex and real-time PCR.
Permingeat HR, Reggiardo MI, Vallejos RH. Permingeat HR, et al. J Agric Food Chem. 2002 Jul 31;50(16):4431-6. doi: 10.1021/jf020081d. J Agric Food Chem. 2002. PMID: 12137456 - Identification and quantification of three genetically modified insect resistant cotton lines using conventional and TaqMan real-time polymerase chain reaction methods.
Yang L, Pan A, Zhang K, Guo J, Yin C, Chen J, Huang C, Zhang D. Yang L, et al. J Agric Food Chem. 2005 Aug 10;53(16):6222-9. doi: 10.1021/jf050095u. J Agric Food Chem. 2005. PMID: 16076097 - Toward metrological traceability for DNA fragment ratios in GM quantification. 1. Effect of DNA extraction methods on the quantitative determination of Bt176 corn by real-time PCR.
Corbisier P, Broothaerts W, Gioria S, Schimmel H, Burns M, Baoutina A, Emslie KR, Furui S, Kurosawa Y, Holden MJ, Kim HH, Lee YM, Kawaharasaki M, Sin D, Wang J. Corbisier P, et al. J Agric Food Chem. 2007 May 2;55(9):3249-57. doi: 10.1021/jf062931l. Epub 2007 Apr 4. J Agric Food Chem. 2007. PMID: 17407305 - [Quantitative PCR in the diagnosis of Leishmania].
Mortarino M, Franceschi A, Mancianti F, Bazzocchi C, Genchi C, Bandi C. Mortarino M, et al. Parassitologia. 2004 Jun;46(1-2):163-7. Parassitologia. 2004. PMID: 15305709 Review. Italian.
Cited by
- Universal probe-based intermediate primer-triggered qPCR (UPIP-qPCR) for SNP genotyping.
Li B, Liu Y, Hao X, Dong J, Chen L, Li H, Wu W, Liu Y, Wang J, Wang Y, Li P. Li B, et al. BMC Genomics. 2021 Nov 24;22(1):850. doi: 10.1186/s12864-021-08148-2. BMC Genomics. 2021. PMID: 34819030 Free PMC article. - Simultaneous digital quantification and fluorescence-based size characterization of massively parallel sequencing libraries.
Laurie MT, Bertout JA, Taylor SD, Burton JN, Shendure JA, Bielas JH. Laurie MT, et al. Biotechniques. 2013 Aug;55(2):61-7. doi: 10.2144/000114063. Biotechniques. 2013. PMID: 23931593 Free PMC article. - A novel universal real-time PCR system using the attached universal duplex probes for quantitative analysis of nucleic acids.
Yang L, Liang W, Jiang L, Li W, Cao W, Wilson ZA, Zhang D. Yang L, et al. BMC Mol Biol. 2008 Jun 4;9:54. doi: 10.1186/1471-2199-9-54. BMC Mol Biol. 2008. PMID: 18522756 Free PMC article. - Digital PCR provides sensitive and absolute calibration for high throughput sequencing.
White RA 3rd, Blainey PC, Fan HC, Quake SR. White RA 3rd, et al. BMC Genomics. 2009 Mar 19;10:116. doi: 10.1186/1471-2164-10-116. BMC Genomics. 2009. PMID: 19298667 Free PMC article. - Potential role of melastatin-related transient receptor potential cation channel subfamily M gene expression in the pathogenesis of urinary bladder cancer.
Ceylan GG, Önalan EE, Kuloğlu T, Aydoğ G, Keleş İ, Tonyali Ş, Ceylan C. Ceylan GG, et al. Oncol Lett. 2016 Dec;12(6):5235-5239. doi: 10.3892/ol.2016.5359. Epub 2016 Nov 7. Oncol Lett. 2016. PMID: 28101241 Free PMC article.
References
- Studer E., Dahinden,I., Luthy,J. and Hubner,P. (1997) Nachweis des genetechnisch veranderten Maximizer-mais mittels der polymerase-kettenreaktion (PCR). Mitt. Gebiete Lebensm. Hyg., 88, 515–524.
- Livak K.J., Flood,S.J.A., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. - PubMed
- Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. - PubMed
- Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–138. - PubMed
- Clegg R.M. (1992) Fluorescent resonance energy transfer and nucleic acids. Methods Enzymol., 211, 353–388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources