Peptide surface display and secretion using two LPXTG-containing surface proteins from Lactobacillus fermentum BR11 - PubMed (original) (raw)
Peptide surface display and secretion using two LPXTG-containing surface proteins from Lactobacillus fermentum BR11
Mark S Turner et al. Appl Environ Microbiol. 2003 Oct.
Erratum in
- Appl Environ Microbiol. 2005 Mar;71(3):1675
Abstract
A locus encoding two repetitive proteins that have LPXTG cell wall anchoring signals from Lactobacillus fermentum BR11 has been identified by using an antiserum raised against whole L. fermentum BR11 cells. The first protein, Rlp, is similar to the Rib surface protein from Streptococcus agalactiae, while the other protein, Mlp, is similar to the mucus binding protein Mub from Lactobacillus reuteri. It was shown that multiple copies of mlp exist in the genome of L. fermentum BR11. Regions of Rlp, Mlp, and the previously characterized surface protein BspA were used to surface display or secrete heterologous peptides in L. fermentum. The peptides tested were 10 amino acids of the human cystic fibrosis transmembrane regulator protein and a six-histidine epitope (His(6)). The BspA promoter and secretion signal were used in combination with the Rlp cell wall sorting signal to express, export, and covalently anchor the heterologous peptides to the cell wall. Detection of the cell surface protein fusions revealed that Rlp was a significantly better surface display vector than BspA despite having lower cellular levels (0.7 mg per liter for the Rlp fusion compared with 4 mg per liter for the BspA fusion). The mlp promoter and encoded secretion signal were used to express and export large (328-kDa at 10 mg per liter) and small (27-kDa at 0.06 mg per liter) amino-terminal fragments of the Mlp protein fused to the His(6) and CFTR peptides or His(6) peptide, respectively. Therefore, these newly described proteins from L. fermentum BR11 have potential as protein production and targeting vectors.
Figures
FIG. 1.
The 16,401-bp DNA region, showing the arrangement of mlp and rlp, and the λ and plasmid clones used to determine the sequence. The letters above the boxes indicate the cell wall sorting (black boxes) and secretion signals of the encoded proteins. The underlined sequences indicate hydrophobic domains. The shaded boxes indicate the encoded amino acid repeat regions, with light and dark boxes indicating the different-sized repeats in Mlp. The lollipop indicates a potential stem-loop structure. The checkered box indicates the DNA region which was amplified by PCR to determine the 3′ end sequence of mlp and to identify multiple mlp genes. The oligonucleotides Mlp-Paw-C and Mlp-C-check (Table 1) were used to amplify this region.
FIG. 2.
Reverse complement of a DNA sequencing trace of the 1.3-kb mlp 3′ PCR fragment (checkered box in Fig. 1) with the Mlp-C-check oligonucleotide. Above the DNA sequence is the encoded amino acid sequence of the Mlp protein, with the LPQTG motif in boldface. Double peaks indicate polymorphic nucleotides, which are included under the assigned sequence. The first polymorphism from the left shows that the encoded amino acid is different in the two Mlp proteins.
FIG. 3.
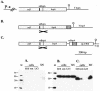
Gene constructs used to integrate into the L. fermentum BR11 bspA locus (top panels) and Western blot detection of fusion proteins in cell extracts and in the supernatant with anti-His5 antibody (bottom panels) for the native bspA genomic locus (A), the BspA-His6-CFTR fusion protein (B), and the His6-CFTR-Rlp fusion protein (C). In the diagrams in the top panels, the bspA upstream promoter (P→), the bspA terminator (T bspA), the rlp terminator (T rlp), and DNA encoding the BspA secretion signal (ssBspA), His6 (grey box), three copies of the CFTR peptide (hatched box), and the cell wall sorting signal of Rlp (_rlp_-CWS) are indicated. The DNA region which is the site of single-crossover homologous recombination into the bspA locus is stippled and marked with a cross. In the bottom panels, the Western blots contained His6 molecular mass markers (with sizes in kilodaltons) (lanes M); cell extracts prepared by boiling in 2× loading dye (lanes SDS), by sonication (lanes son), and with 5 M LiCl (lanes LiCl); and precipitated supernatant fractions (lanes SN). The amounts of cells or medium loaded in each lane are equivalent to 500 μl (lanes SDS), 50 μl (lanes son), 160 μl (lanes LiCl), and 625 μl (lanes SN) of culture.
FIG. 4.
Gene constructs used to integrate into the L. fermentum BR11 mlp locus (top panels) and Western blot detection of fusion proteins in cell extracts and in the supernatant with anti-His5 antibody (bottom panels) for the Mlp-His6 fusion protein (A) and the Mlp-His6-CFTR fusion protein (B). In the diagrams in the top panels, the bspA terminator (T bspA) and DNA encoding the Mlp secretion signal (ssMlp), His6 (grey box), and three copies of the CFTR peptide (hatched box) are indicated. The DNA region which is the site of single-crossover homologous recombination into the mlp locus is stippled and marked with a cross. Due to the large size of mlp, the full length of the gene could not be shown. In the bottom panels, the Western blots contained His6 molecular mass markers (with sizes in kilodaltons) (lanes M); cell extracts prepared by boiling in 2× loading dye (lanes SDS), by sonication (lanes son), and with 5 M LiCl (lanes LiCl); and precipitated supernatant fractions (lanes SN). The amounts of cells or medium loaded in each lane are the equivalent to 500 μl (lanes SDS), 50 μl (lanes son), 160 μl (lanes LiCl), and 625 μl (lanes SN) of culture.
Similar articles
- Identification and characterization of the novel LysM domain-containing surface protein Sep from Lactobacillus fermentum BR11 and its use as a peptide fusion partner in Lactobacillus and Lactococcus.
Turner MS, Hafner LM, Walsh T, Giffard PM. Turner MS, et al. Appl Environ Microbiol. 2004 Jun;70(6):3673-80. doi: 10.1128/AEM.70.6.3673-3680.2004. Appl Environ Microbiol. 2004. PMID: 15184172 Free PMC article. - Identification and characterization of a basic cell surface-located protein from Lactobacillus fermentum BR11.
Turner MS, Timms P, Hafner LM, Giffard PM. Turner MS, et al. J Bacteriol. 1997 May;179(10):3310-6. doi: 10.1128/jb.179.10.3310-3316.1997. J Bacteriol. 1997. PMID: 9150229 Free PMC article. - The bspA locus of Lactobacillus fermentum BR11 encodes an L-cystine uptake system.
Turner MS, Woodberry T, Hafner LM, Giffard PM. Turner MS, et al. J Bacteriol. 1999 Apr;181(7):2192-8. doi: 10.1128/JB.181.7.2192-2198.1999. J Bacteriol. 1999. PMID: 10094698 Free PMC article. - Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications.
Michon C, Langella P, Eijsink VG, Mathiesen G, Chatel JM. Michon C, et al. Microb Cell Fact. 2016 May 3;15:70. doi: 10.1186/s12934-016-0468-9. Microb Cell Fact. 2016. PMID: 27142045 Free PMC article. Review. - Peptide display on bacterial flagella: principles and applications.
Westerlund-Wikström B. Westerlund-Wikström B. Int J Med Microbiol. 2000 Jul;290(3):223-30. doi: 10.1016/S1438-4221(00)80119-8. Int J Med Microbiol. 2000. PMID: 10959724 Review.
Cited by
- Putative Adhesion Factors in Vaginal Lactobacillus gasseri DSM 14869: Functional Characterization.
Zeng Z, Zuo F, Marcotte H. Zeng Z, et al. Appl Environ Microbiol. 2019 Sep 17;85(19):e00800-19. doi: 10.1128/AEM.00800-19. Print 2019 Oct 1. Appl Environ Microbiol. 2019. PMID: 31420338 Free PMC article. - Display of a β-mannanase and a chitosanase on the cell surface of Lactobacillus plantarum towards the development of whole-cell biocatalysts.
Nguyen HM, Mathiesen G, Stelzer EM, Pham ML, Kuczkowska K, Mackenzie A, Agger JW, Eijsink VG, Yamabhai M, Peterbauer CK, Haltrich D, Nguyen TH. Nguyen HM, et al. Microb Cell Fact. 2016 Oct 4;15(1):169. doi: 10.1186/s12934-016-0570-z. Microb Cell Fact. 2016. PMID: 27716231 Free PMC article. - Formation of In Vitro Mixed-Species Biofilms by Lactobacillus pentosus and Yeasts Isolated from Spanish-Style Green Table Olive Fermentations.
León-Romero Á, Domínguez-Manzano J, Garrido-Fernández A, Arroyo-López FN, Jiménez-Díaz R. León-Romero Á, et al. Appl Environ Microbiol. 2015 Nov 13;82(2):689-95. doi: 10.1128/AEM.02727-15. Print 2016 Jan 15. Appl Environ Microbiol. 2015. PMID: 26567305 Free PMC article. - High quality draft genome of Lactobacillus kunkeei EFB6, isolated from a German European foulbrood outbreak of honeybees.
Djukic M, Poehlein A, Strauß J, Tann FJ, Leimbach A, Hoppert M, Daniel R. Djukic M, et al. Stand Genomic Sci. 2015 Feb 19;10:16. doi: 10.1186/1944-3277-10-16. eCollection 2015. Stand Genomic Sci. 2015. PMID: 26203329 Free PMC article. - Saccharomyces cerevisiae transcriptional reprograming due to bacterial contamination during industrial scale bioethanol production.
Carvalho-Netto OV, Carazzolle MF, Mofatto LS, Teixeira PJ, Noronha MF, Calderón LA, Mieczkowski PA, Argueso JL, Pereira GA. Carvalho-Netto OV, et al. Microb Cell Fact. 2015 Jan 30;14:13. doi: 10.1186/s12934-015-0196-6. Microb Cell Fact. 2015. PMID: 25633848 Free PMC article.
References
- Antikainen, J., L. Anton, J. Sillanpaa, and T. K. Korhonen. 2002. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46:381-394. - PubMed
- Beninati, C., M. R. Oggioni, M. Boccanera, M. R. Spinosa, T. Maggi, S. Conti, W. Magliani, F. De Bernardis, G. Teti, A. Cassone, G. Pozzi, and L. Polonelli. 2000. Therapy of mucosal candidiasis by expression of an anti-idiotype in human commensal bacteria. Nat. Biotechnol. 18:1060-1064. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources