Single-cell enumeration of an uncultivated TM7 subgroup in the human subgingival crevice - PubMed (original) (raw)
Single-cell enumeration of an uncultivated TM7 subgroup in the human subgingival crevice
Cleber C Ouverney et al. Appl Environ Microbiol. 2003 Oct.
Abstract
Specific oligonucleotide hybridization conditions were established for single-cell enumeration of uncultivated TM7 and IO25 bacteria by using clones expressing heterologous 16S rRNA. In situ analysis of human subgingival crevice specimens revealed that a greater proportion of samples from sites of chronic periodontitis than from healthy sites contained TM7 subgroup IO25. In addition, IO25 bacterial cells from periodontitis site samples were more abundant and fourfold longer than IO25 cells from healthy site samples.
Figures
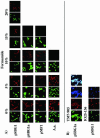
FIG. 1.
(A) FISH stringency tests with clones pSBG1, pSBG1a, pMF1, and A. aquaticus and probe TM7-905 at 0 to 20% formamide concentrations. All green panels refer to the use of the nonspecific YO-PRO-1 DNA stain, and red panels refer to the use of the Cy3-labeled TM7-905-specific probe. (B) Staining with TM7-905-Cy3 (10% formamide; clone pSBG1a) or IO25-136-Cy3 (20% formamide; pSBG1), demonstrating, in addition to YO-PRO-1 (green) and probe-Cy3 (red) labels, the positive-control Bac338-Cy5 (blue) probe. The last TM7-905 panel shows colocalization of the three stains with heterologous 16S rRNA at the cell poles. Scale bar = 5 μm for all panels.
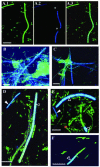
FIG. 2.
FISH on dental plaque samples from human subgingival sites of moderate chronic periodontitis (A to E) and from healthy sites (F). (A.1 to A.3) Staining with YO-PRO-1 (green) general stain (A.1) and TM7-905-Cy5 (blue) (A.2) and colocalization of these two stains (A.3). Asterisks, two segments of the TM7 filament that are not labeled by the probe. In all other panels, YO-PRO-1 and probe signals are merged as in panel A.3. (B and C) TM7-905 probe and YO-PRO-1-labeled cells (blue-green); (D to F) TM7-905 probe-labeled cells (solid arrowhead, dark blue) plus IO25-136 probe-labeled cells (open arrowhead, light blue) and cells labeled with YO-PRO-1 alone (green). Scale bar = 5 μm in all panels.
Similar articles
- Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease.
Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Brinig MM, et al. Appl Environ Microbiol. 2003 Mar;69(3):1687-94. doi: 10.1128/AEM.69.3.1687-1694.2003. Appl Environ Microbiol. 2003. PMID: 12620860 Free PMC article. - RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms.
Teles FR, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. Teles FR, et al. Mol Oral Microbiol. 2011 Apr;26(2):127-39. doi: 10.1111/j.2041-1014.2010.00603.x. Epub 2011 Jan 27. Mol Oral Microbiol. 2011. PMID: 21375703 Free PMC article. - Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets.
de Lillo A, Ashley FP, Palmer RM, Munson MA, Kyriacou L, Weightman AJ, Wade WG. de Lillo A, et al. Oral Microbiol Immunol. 2006 Feb;21(1):61-8. doi: 10.1111/j.1399-302X.2005.00255.x. Oral Microbiol Immunol. 2006. PMID: 16390343 - Does routine analysis of subgingival microbiota in periodontitis contribute to patient benefit?
Fernandez y Mostajo M, Zaura E, Crielaard W, Beertsen W. Fernandez y Mostajo M, et al. Eur J Oral Sci. 2011 Aug;119(4):259-64. doi: 10.1111/j.1600-0722.2011.00828.x. Epub 2011 May 17. Eur J Oral Sci. 2011. PMID: 21726285 Review. - Subgingival biofilm structure.
Zijnge V, Ammann T, Thurnheer T, Gmür R. Zijnge V, et al. Front Oral Biol. 2012;15:1-16. doi: 10.1159/000329667. Epub 2011 Nov 11. Front Oral Biol. 2012. PMID: 22142954 Review.
Cited by
- Genetic manipulation of Patescibacteria provides mechanistic insights into microbial dark matter and the epibiotic lifestyle.
Wang Y, Gallagher LA, Andrade PA, Liu A, Humphreys IR, Turkarslan S, Cutler KJ, Arrieta-Ortiz ML, Li Y, Radey MC, McLean JS, Cong Q, Baker D, Baliga NS, Peterson SB, Mougous JD. Wang Y, et al. Cell. 2023 Oct 26;186(22):4803-4817.e13. doi: 10.1016/j.cell.2023.08.017. Epub 2023 Sep 7. Cell. 2023. PMID: 37683634 Free PMC article. - Molecular profiling of oral microbiota in jawbone samples of bisphosphonate-related osteonecrosis of the jaw.
Wei X, Pushalkar S, Estilo C, Wong C, Farooki A, Fornier M, Bohle G, Huryn J, Li Y, Doty S, Saxena D. Wei X, et al. Oral Dis. 2012 Sep;18(6):602-12. doi: 10.1111/j.1601-0825.2012.01916.x. Epub 2012 Mar 23. Oral Dis. 2012. PMID: 22443347 Free PMC article. - Developing animal models for polymicrobial diseases.
Bakaletz LO. Bakaletz LO. Nat Rev Microbiol. 2004 Jul;2(7):552-68. doi: 10.1038/nrmicro928. Nat Rev Microbiol. 2004. PMID: 15197391 Free PMC article. Review. - Molecular microbial diagnosis.
Paster BJ, Dewhirst FE. Paster BJ, et al. Periodontol 2000. 2009;51:38-44. doi: 10.1111/j.1600-0757.2009.00316.x. Periodontol 2000. 2009. PMID: 19878468 Free PMC article. Review. No abstract available. - NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis.
Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. Elinav E, et al. Cell. 2011 May 27;145(5):745-57. doi: 10.1016/j.cell.2011.04.022. Epub 2011 May 12. Cell. 2011. PMID: 21565393 Free PMC article.
References
- Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. - PubMed
- Hugenholtz, P., G. W. Tyson, and L. L. Blackall. 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179:29-42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE013541/DE/NIDCR NIH HHS/United States
- T32 AI007328/AI/NIAID NIH HHS/United States
- 5 T32 AI07328/AI/NIAID NIH HHS/United States
- R01 DE13541/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources