Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import - PubMed (original) (raw)
Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import
Yoshiyuki Matsuura et al. EMBO J. 2003.
Abstract
The yeast nucleoporin Nup2p is associated primarily with the nuclear basket of nuclear pore complexes and is required for efficient importin-alpha:beta-mediated nuclear protein import as well as efficient nuclear export of Kap60p/importin-alpha. Residues 1-51 of Nup2p bind tightly to Kap60p and are required for Nup2p function in vivo. We have determined the 2.6 A resolution crystal structure of a complex between this region of Nup2p and the armadillo repeat domain of Kap60p. Nup2p binds along the inner concave groove of Kap60p, but its interaction interface is different from that employed for nuclear localization signal (NLS) recognition although there is some overlap between them. Nup2p binds Kap60p more strongly than NLSs and accelerates release of NLSs from Kap60p. Nup2p itself is released from Kap60p by Cse1p:RanGTP only in the presence of the importin-beta binding (IBB) domain of Kap60p. These data indicate that Nup2p increases the overall rate of nuclear trafficking by coordinating nuclear import termination and importin recycling as a concerted process.
Figures
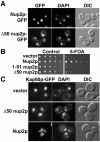
Fig. 1. In vivo functional analysis of NUP2. (A) Nup2p and Δ50Nup2p expressed as C-terminal GFP fusions under the control of NUP2 promoter in Δ_nup2_ yeast cells show nuclear envelope localization as visualized by GFP fluorescence. Corresponding DIC and DAPI images are shown. (B) Δ_nup2 srp1-31_ yeast cells maintained by a plasmid encoding Kap60p and expressing either full-length Nup2p, 1–51 Nup2p or Δ50Nup2p were spotted onto control plates lacking uracil or 5-FOA plates. 5-FOA eliminates the URA3 maintenance plasmid encoding Kap60p. (C) The Nup2p N-terminus promotes docking of Kap60p to the nuclear envelope and is required for efficient recycling of Kap60p to the cytoplasm. Kap60p–GFP was integrated at the endogenous SRP1 locus of Δ_nup2_ yeast cells. The cells were then transformed with plasmids encoding either full-length Nup2p, Δ50Nup2p, or vector alone, and Kap60p–GFP was visualized by GFP fluorescence. Corresponding DIC and DAPI images are shown.
Fig. 2. Structure of residues 1–51 of Nup2p bound to Kap60Δ. (A) Overview of the Kap60Δ:Nup2N complex. The _F_o – _F_c omit map corresponding to the Nup2p fragment contoured at 2.5 σ and the refined model of Nup2p (residues 36–51) are superimposed. (B) Overlay of Nup2p with NLS peptides. The coordinates of Kap60Δ bound to Nup2N (blue) and the SV40 (purple), c-myc (yellow), and nucleoplasmin (green) NLSs (Conti et al., 1998; Conti and Kuriyan, 2000) were superimposed, then removed to show the relative positions of each ligand. Orientation as in (A). (A and B) were prepared with Bobscript (Esnouf, 1997) and Raster3D (Merritt and Bacon, 1997). (C) Molecular surface of Kap60Δ coloured by electrostatic potential shaded from –13 kT/e (red) to +13 kT/e (blue) (calculated with Nup2N removed using GRASP; Nicholls et al., 1991) shows acidic pockets and a nonpolar surface that recognize Nup2p. Nup2p residues are bold.
Fig. 3. Schematic comparison of the interactions of Kap60Δ with (A) Nup2p, (B) IBB domain (based on mouse importin α; Kobe, 1999), (C) monopartite NLS (Conti et al., 1998), and (D) bipartite NLS (Conti and Kuriyan, 2000).
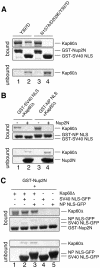
Fig. 4. Nup2p–Kap60p interactions are different from NLS interactions and Nup2p competes with NLSs. (A) N157A/D203K mutations in the major NLS binding site of Kap60Δ abolish SV40 NLS binding but not Nup2N binding. GST–Nup2N (10 µg; lanes 1 and 3) or GST–SV40 NLS (10 µg; lanes 2 and 4) was treated with 15 µg of Kap60Δ. (B) Nup2N competes both monopartite and bipartite NLSs from Kap60Δ. Beads containing 4.5 µg GST–SV40 NLS or GST–nucleoplasmin (NP) NLS were treated with Kap60Δ (10 µg) which remained mainly bound after washing with binding buffer but was subsequently removed by 30 µM His/S–Nup2N. (C) NLS-cargoes do not effectively compete Nup2N from Kap60Δ and only bind weakly in the presence of Nup2N. GST––Nup2N (4.5 µg) was treated with Kap60Δ (10 µg) ± 30 µM NLS–GFP (lanes 1–3) or 30 µM NLS–GFP alone (lanes 4 and 5).
Fig. 5. Nup2N accelerates NLS dissociation. (A) Emission profiles of 0.2 µM BFP–Kap60Δ (line 1), 0.18 µM SV40 NLS–GFP (line 2), 0.2 µM BFP–Kap60Δ and 0.18 µM SV40 NLS–GFP (line 3), 0.2 µM BFP–Kap60Δ, 0.18 µM SV40 NLS–GFP and 2 µM Nup2N (line 4). Excitation was at 360 nm (BFP excitation peak). (B) Nup2N increases the off-rate of SV40 NLS. 0.2 µM BFP–Kap60Δ and 0.18 µM SV40 NLS–GFP alone (line 1), or with 4 µM Kap60Δ (line 2), or 2 µM Nup2N (line 3) added. The line 3 trace is expanded on the right. (C) Nup2N increases the off-rate of nucleoplasmin (NP) NLS. 0.2 µM BFP–Kap60Δ and 0.18 µM NP NLS–GFP alone (line 1), or with 4 µM Kap60Δ (line 2), or 2 µM Nup2N (line 3) added. The line 3 trace is expanded on the right.
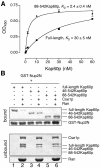
Fig. 6. Competition with the IBB domain and Cse1p/RanGTP. (A) Kap60p (filled squares) binds more weakly to Nup2N than Kap60Δ (filled triangles). Each data point was performed in duplicate and error bars represent SEM. (B) Cse1p:RanGTP competes Kap60p from Nup2N when the IBB domain is present. GST–Nup2N (10 µg) was treated with 3 µM Kap60p ± Cse1p (15 µg):Ran (30 µg) in binding buffer supplemented with 5 mM Mg(OAc)2. Two bands marked with * in the unbound fractions are copurifying bacterial proteins present in Cse1p. Full-length Kap60p can be seen between these two bands in lane 2.
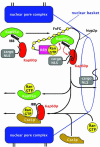
Fig. 7. Proposed mechanism by which Nup2p orchestrates dissociation of the import complex and recycling of Kap95p and Kap60p. Docking of the Kap60p:Kap95p:NLS-cargo complex to Nup2p at the nuclear basket triggers cooperative binding/dissociation interactions leading to release of the NLS-cargo into the nucleoplasm concomitant with release of export complexes from nuclear basket. The Nup2p N-terminus displaces NLS-cargo from Kap60p. By transferring RanGTP to Kap95p and recruiting Cse1p and RanGTP to Kap60p, Nup2p further facilitates NLS-cargo release as well as facilitating formation of export complexes. RanGTP binding to Kap95p dissociates it from the FxFG repeats of Nup2p and the IBB domain of Kap60p. Cse1p and RanGTP then bind to Kap60p so that Cse1p:RanGTP stabilizes the interaction between the IBB domain and the Arm repeat domain of Kap60p, ensuring both NLS and Nup2p dissociation. Thus, Nup2p orchestrates both the termination of nuclear protein import and the initiation of the nuclear export of cargo-free importins through interactions involving the Kap60p IBB domain, RanGTP and Cse1p.
Similar articles
- Accelerating the rate of disassembly of karyopherin.cargo complexes.
Gilchrist D, Mykytka B, Rexach M. Gilchrist D, et al. J Biol Chem. 2002 May 17;277(20):18161-72. doi: 10.1074/jbc.M112306200. Epub 2002 Feb 26. J Biol Chem. 2002. PMID: 11867631 - Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm.
Gilchrist D, Rexach M. Gilchrist D, et al. J Biol Chem. 2003 Dec 19;278(51):51937-49. doi: 10.1074/jbc.M307371200. Epub 2003 Sep 26. J Biol Chem. 2003. PMID: 14514698 - Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha.
Solsbacher J, Maurer P, Vogel F, Schlenstedt G. Solsbacher J, et al. Mol Cell Biol. 2000 Nov;20(22):8468-79. doi: 10.1128/MCB.20.22.8468-8479.2000. Mol Cell Biol. 2000. PMID: 11046143 Free PMC article. - The importin β binding domain as a master regulator of nucleocytoplasmic transport.
Lott K, Cingolani G. Lott K, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review. - Karyopherins and nuclear import.
Chook YM, Blobel G. Chook YM, et al. Curr Opin Struct Biol. 2001 Dec;11(6):703-15. doi: 10.1016/s0959-440x(01)00264-0. Curr Opin Struct Biol. 2001. PMID: 11751052 Review.
Cited by
- Managing free-energy barriers in nuclear pore transport.
Nielsen B, Jeppesen C, Ipsen JH. Nielsen B, et al. J Biol Phys. 2006 Nov;32(5):465-72. doi: 10.1007/s10867-006-9029-5. Epub 2006 Dec 19. J Biol Phys. 2006. PMID: 19669451 Free PMC article. - Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex.
Fiserova J, Richards SA, Wente SR, Goldberg MW. Fiserova J, et al. J Cell Sci. 2010 Aug 15;123(Pt 16):2773-80. doi: 10.1242/jcs.070730. Epub 2010 Jul 20. J Cell Sci. 2010. PMID: 20647373 Free PMC article. - Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo.
Fox AM, Ciziene D, McLaughlin SH, Stewart M. Fox AM, et al. J Biol Chem. 2011 Aug 19;286(33):29325-29335. doi: 10.1074/jbc.M111.245092. Epub 2011 Jun 27. J Biol Chem. 2011. PMID: 21708948 Free PMC article. - The Nucleoporin Nup2 Contains a Meiotic-Autonomous Region that Promotes the Dynamic Chromosome Events of Meiosis.
Chu DB, Gromova T, Newman TAC, Burgess SM. Chu DB, et al. Genetics. 2017 Jul;206(3):1319-1337. doi: 10.1534/genetics.116.194555. Epub 2017 Apr 28. Genetics. 2017. PMID: 28455351 Free PMC article. - Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein.
Nakada R, Hirano H, Matsuura Y. Nakada R, et al. Sci Rep. 2015 Oct 12;5:15055. doi: 10.1038/srep15055. Sci Rep. 2015. PMID: 26456934 Free PMC article.
References
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. - PubMed
- Bayliss R., Littlewood,T., Strawn,L.A., Wente,S.R. and Stewart,M. (2002) GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J. Biol. Chem., 277, 50597–50606. - PubMed
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. - PubMed
- Booth J.W., Belanger,K.D., Sannella,M.I. and Davis,L.I. (1999) The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J. Biol. Chem., 274, 32360–32367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases