Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions - PubMed (original) (raw)
Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions
Femke Simmer et al. PLoS Biol. 2003 Oct.
Abstract
RNA-mediated interference (RNAi) is a method to inhibit gene function by introduction of double-stranded RNA (dsRNA). Recently, an RNAi library was constructed that consists of bacterial clones expressing dsRNA, corresponding to nearly 90% of the 19,427 predicted genes of C. elegans. Feeding of this RNAi library to the standard wild-type laboratory strain Bristol N2 detected phenotypes for approximately 10% of the corresponding genes. To increase the number of genes for which a loss-of-function phenotype can be detected, we undertook a genome-wide RNAi screen using the rrf-3 mutant strain, which we found to be hypersensitive to RNAi. Feeding of the RNAi library to rrf-3 mutants resulted in additional loss-of-function phenotypes for 393 genes, increasing the number of genes with a phenotype by 23%. These additional phenotypes are distributed over different phenotypic classes. We also studied interexperimental variability in RNAi results and found persistent levels of false negatives. In addition, we used the RNAi phenotypes obtained with the genome-wide screens to systematically clone seven existing genetic mutants with visible phenotypes. The genome-wide RNAi screen using rrf-3 significantly increased the functional data on the C. elegans genome. The resulting dataset will be valuable in conjunction with other functional genomics approaches, as well as in other model organisms.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
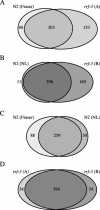
Figure 1. Comparison of Different RNAi Experiments of Chromosome I Using Wild-Type Bristol N2 and rrf-3
Differences between different laboratories or investigators and between experiments done within the same laboratory and by the same investigators are observed. Ovals represent the amount of bacterial clones that gave an RNAi phenotype in an experiment. Areas that overlap represent clones for which in both experiments an RNAi phenotype was detected. Differences and overlap between an RNAi experiment done with the rrf-3 mutant strain and the data obtained by Fraser et al. (2000) done with the standard laboratory strain, Bristol N2 (A); N2 and rrf-3 tested at the same time within our laboratory (B); experiments done with N2 in two different laboratories: this study (‘NL') and Fraser et al. (2000) (C); two experiments done with the same strain, rrf-3, within our laboratory (D).
Similar articles
- Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi.
Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Simmer F, et al. Curr Biol. 2002 Aug 6;12(15):1317-9. doi: 10.1016/s0960-9822(02)01041-2. Curr Biol. 2002. PMID: 12176360 - Uncover genetic interactions in Caenorhabditis elegans by RNA interference.
Fortunato A, Fraser AG. Fortunato A, et al. Biosci Rep. 2005 Oct-Dec;25(5-6):299-307. doi: 10.1007/s10540-005-2892-7. Biosci Rep. 2005. PMID: 16307378 Review. - Functional genomic approaches in C. elegans.
Lamitina T. Lamitina T. Methods Mol Biol. 2006;351:127-38. doi: 10.1385/1-59745-151-7:127. Methods Mol Biol. 2006. PMID: 16988431 Review. - Functional genomic analysis of C. elegans chromosome I by systematic RNA interference.
Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Fraser AG, et al. Nature. 2000 Nov 16;408(6810):325-30. doi: 10.1038/35042517. Nature. 2000. PMID: 11099033 - Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference.
Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, Fraser AG. Lehner B, et al. Genome Biol. 2006;7(1):R4. doi: 10.1186/gb-2006-7-1-r4. Epub 2006 Jan 20. Genome Biol. 2006. PMID: 16507136 Free PMC article.
Cited by
- The atypical calpains: evolutionary analyses and roles in Caenorhabditis elegans cellular degeneration.
Joyce PI, Satija R, Chen M, Kuwabara PE. Joyce PI, et al. PLoS Genet. 2012;8(3):e1002602. doi: 10.1371/journal.pgen.1002602. Epub 2012 Mar 29. PLoS Genet. 2012. PMID: 22479198 Free PMC article. - Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex.
Richard M, Boulin T, Robert VJ, Richmond JE, Bessereau JL. Richard M, et al. Proc Natl Acad Sci U S A. 2013 Mar 12;110(11):E1055-63. doi: 10.1073/pnas.1216154110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431131 Free PMC article. - Caenorhabditis elegans screen reveals role of PAR-5 in RAB-11-recycling endosome positioning and apicobasal cell polarity.
Winter JF, Höpfner S, Korn K, Farnung BO, Bradshaw CR, Marsico G, Volkmer M, Habermann B, Zerial M. Winter JF, et al. Nat Cell Biol. 2012 May 27;14(7):666-76. doi: 10.1038/ncb2508. Nat Cell Biol. 2012. PMID: 22634595 - Nucleolar localization of GLTSCR2/PICT-1 is mediated by multiple unique nucleolar localization sequences.
Kalt I, Levy A, Borodianskiy-Shteinberg T, Sarid R. Kalt I, et al. PLoS One. 2012;7(1):e30825. doi: 10.1371/journal.pone.0030825. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22292050 Free PMC article. - Molecular and biochemical analysis of the beta class carbonic anhydrases in Caenorhabditis elegans.
Fasseas MK, Tsikou D, Flemetakis E, Katinakis P. Fasseas MK, et al. Mol Biol Rep. 2010 Jul;37(6):2941-50. doi: 10.1007/s11033-009-9857-z. Epub 2009 Oct 9. Mol Biol Rep. 2010. PMID: 19816790
References
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. - PubMed
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. - PubMed
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. - PubMed
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. - PubMed
- Gönczy P, Echeverri C, Oegema K, Coulson A, Jones SJM, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources