Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action - PubMed (original) (raw)
Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action
Inês Barroso et al. PLoS Biol. 2003 Oct.
Erratum in
- PLoS Biol. 2003 Dec;1(3):445
Abstract
Type 2 diabetes is an increasingly common, serious metabolic disorder with a substantial inherited component. It is characterised by defects in both insulin secretion and action. Progress in identification of specific genetic variants predisposing to the disease has been limited. To complement ongoing positional cloning efforts, we have undertaken a large-scale candidate gene association study. We examined 152 SNPs in 71 candidate genes for association with diabetes status and related phenotypes in 2,134 Caucasians in a case-control study and an independent quantitative trait (QT) cohort in the United Kingdom. Polymorphisms in five of 15 genes (33%) encoding molecules known to primarily influence pancreatic beta-cell function-ABCC8 (sulphonylurea receptor), KCNJ11 (KIR6.2), SLC2A2 (GLUT2), HNF4A (HNF4alpha), and INS (insulin)-significantly altered disease risk, and in three genes, the risk allele, haplotype, or both had a biologically consistent effect on a relevant physiological trait in the QT study. We examined 35 genes predicted to have their major influence on insulin action, and three (9%)-INSR, PIK3R1, and SOS1-showed significant associations with diabetes. These results confirm the genetic complexity of Type 2 diabetes and provide evidence that common variants in genes influencing pancreatic beta-cell function may make a significant contribution to the inherited component of this disease. This study additionally demonstrates that the systematic examination of panels of biological candidate genes in large, well-characterised populations can be an effective complement to positional cloning approaches. The absence of large single-gene effects and the detection of multiple small effects accentuate the need for the study of larger populations in order to reliably identify the size of effect we now expect for complex diseases.
Conflict of interest statement
Portions of the research were supported by Incyte Corporation (formerly Incyte Genomics). This collaboration provided access to certain technologies and scientific expertise for basic research in the genetics of diabetes in return for possible shared intellectual property. The work described here is not subject to any patent filings or intellectual property protection and restrictions from free use of the information. IB and AJS were employees of Incyte at the time the research took place.
Figures
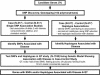
Figure 1. Study Design
Candidate genes were selected based on known or putative function. A de novo polymorphism discovery step was undertaken to identify novel variants for association studies. We selected 152 SNPs and tested them in a case-control study and a QT study. Association analysis with Type 2 diabetes was done for SNPs and haplotypes under multiple genetic models. Only SNPs and haplotypes associated with disease were evaluated for association with five diabetes-related QTs under the same model in the QT study.
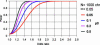
Figure 2. Power Calculations
Power of the current Cambridgeshire Case-Control Study to detect associations with risk allele of varying frequencies and with a Type 1 error rate of 5%. Abbreviations: p0, frequency of the predisposing allele; chr, number of chromosomes. Graphs were plotted with the PS power and sample-size program (available at
http://www.mc.vanderbilt.edu/prevmed/ps
; DuPont and Plummer 1997).
Figure 3. Genes with Haplotypes Associated with Type 2 Diabetes
Genomic organization with exons (black boxes or vertical lines) and with genotyped SNPs and SNPs utilised in the haplotype reconstructions (in blue boxes) is shown. The most common haplotypes with population prevalence greater than 5% in the control population are shown, and the measure of LD (r2) is shown for a subset of the SNPs. (A) ABCC8–KCNJ11. (B) HNF4A. (C) INSR.
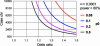
Figure 4. Size of Case-Control Study Required to Detect Small Risk Effects
The number is shown of the case chromosomes (assuming an equal number of control chromosomes) required to attain 80% power to detect associations with the OR varying between 1.0 and 1.5 and with a Type 1 error rate of 0.01%. Abbreviations: p0, frequency of the predisposing allele; chr, number of chromosomes. Graphs were plotted with the PS power and sample-size program (DuPont and Plummer 1997).
Similar articles
- Physical activity modifies the effect of SNPs in the SLC2A2 (GLUT2) and ABCC8 (SUR1) genes on the risk of developing type 2 diabetes.
Kilpeläinen TO, Lakka TA, Laaksonen DE, Laukkanen O, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Aunola S, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Tuomilehto J, Uusitupa M, Laakso M; Finnish Diabetes Prevention Study Group. Kilpeläinen TO, et al. Physiol Genomics. 2007 Oct 22;31(2):264-72. doi: 10.1152/physiolgenomics.00036.2007. Epub 2007 Jul 17. Physiol Genomics. 2007. PMID: 17636114 Clinical Trial. - Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region.
Florez JC, Burtt N, de Bakker PI, Almgren P, Tuomi T, Holmkvist J, Gaudet D, Hudson TJ, Schaffner SF, Daly MJ, Hirschhorn JN, Groop L, Altshuler D. Florez JC, et al. Diabetes. 2004 May;53(5):1360-8. doi: 10.2337/diabetes.53.5.1360. Diabetes. 2004. PMID: 15111507 - Effects of single nucleotide polymorphisms in K(ATP) channel genes on type 2 diabetes in a Turkish population.
Gonen MS, Arikoglu H, Erkoc Kaya D, Ozdemir H, Ipekci SH, Arslan A, Kayis SA, Gogebakan B. Gonen MS, et al. Arch Med Res. 2012 May;43(4):317-23. doi: 10.1016/j.arcmed.2012.06.001. Epub 2012 Jun 13. Arch Med Res. 2012. PMID: 22704848 - Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism.
Flanagan SE, Clauin S, Bellanné-Chantelot C, de Lonlay P, Harries LW, Gloyn AL, Ellard S. Flanagan SE, et al. Hum Mutat. 2009 Feb;30(2):170-80. doi: 10.1002/humu.20838. Hum Mutat. 2009. PMID: 18767144 Review.
Cited by
- SNP-Based Genetic Risk Score Modeling Suggests No Increased Genetic Susceptibility of the Roma Population to Type 2 Diabetes Mellitus.
Werissa NA, Piko P, Fiatal S, Kosa Z, Sandor J, Adany R. Werissa NA, et al. Genes (Basel). 2019 Nov 19;10(11):942. doi: 10.3390/genes10110942. Genes (Basel). 2019. PMID: 31752367 Free PMC article. - PPARD rs2016520 (T/C) and NOS1AP rs12742393 (A/C) polymorphisms affect therapeutic efficacy of nateglinide in Chinese patients with type 2 diabetes mellitus.
Wang T, Song JF, Zhou XY, Li CL, Yin XX, Lu Q. Wang T, et al. BMC Med Genomics. 2021 Nov 12;14(1):267. doi: 10.1186/s12920-021-01108-5. BMC Med Genomics. 2021. PMID: 34772419 Free PMC article. - The role of membrane glycoprotein plasma cell antigen 1/ectonucleotide pyrophosphatase phosphodiesterase 1 in the pathogenesis of insulin resistance and related abnormalities.
Goldfine ID, Maddux BA, Youngren JF, Reaven G, Accili D, Trischitta V, Vigneri R, Frittitta L. Goldfine ID, et al. Endocr Rev. 2008 Feb;29(1):62-75. doi: 10.1210/er.2007-0004. Epub 2008 Jan 16. Endocr Rev. 2008. PMID: 18199690 Free PMC article. Review. - Mutations in SLC2A2 gene reveal hGLUT2 function in pancreatic β cell development.
Michau A, Guillemain G, Grosfeld A, Vuillaumier-Barrot S, Grand T, Keck M, L'Hoste S, Chateau D, Serradas P, Teulon J, De Lonlay P, Scharfmann R, Brot-Laroche E, Leturque A, Le Gall M. Michau A, et al. J Biol Chem. 2013 Oct 25;288(43):31080-92. doi: 10.1074/jbc.M113.469189. Epub 2013 Aug 28. J Biol Chem. 2013. PMID: 23986439 Free PMC article. - Genome-wide meta-analysis of genetic susceptible genes for Type 2 Diabetes.
Hale PJ, López-Yunez AM, Chen JY. Hale PJ, et al. BMC Syst Biol. 2012;6 Suppl 3(Suppl 3):S16. doi: 10.1186/1752-0509-6-S3-S16. Epub 2012 Dec 17. BMC Syst Biol. 2012. PMID: 23281828 Free PMC article.
References
- Alpha B, Cox L, Crowther N, Clark PM, Hales CN. Sensitive amplified immunoenzymometric assays (IEMA) for human insulin and intact proinsulin. Eur J Clin Chem Clin Biochem. 1992;30:27–32. - PubMed
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. - PubMed
- Argyrokastritis A, Kamakari S, Kapsetaki M, Kritis A, Talianidis I, et al. Human hepatocyte nuclear factor-4 (hHNF-4) gene maps to 20q12–q13.1 between PLCG1 and D20S17. Hum Genet. 1997;99:233–236. - PubMed
- Baier LJ, Wiedrich C, Hanson RL, Bogardus C. Variant in the regulatory subunit of phosphatidylinositol 3-kinase (p85α): Preliminary evidence indicates a potential role of this variant in the acute insulin response and type 2 diabetes in Pima women. Diabetes. 1998;47:973–975. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous