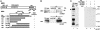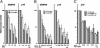Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin - PubMed (original) (raw)
Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin
Sugiko Watanabe et al. Proc Natl Acad Sci U S A. 2003.
Abstract
The methyl-CpG dinucleotide containing a symmetrical 5-methylcytosine (mC) is involved in gene regulation and genome stability. We report here that methylation-mediated transcriptional repressor methylated DNA-binding domain 1 (MBD1) interacts with methylpurine-DNA glycosylase (MPG), which excises damaged bases from substrate DNA. MPG itself actively represses transcription and has a synergistic effect on gene silencing together with MBD1. Chromatin immunoprecipitation analysis reveals the molecular movement of MBD1 and MPG in vivo: (i) The MBD1-MPG complex normally exists on the methylated gene promoter; (ii) treatment of cells with alkylating agent methylmethanesulfonate (MMS) induces the dissociation of MBD1 from the methylated promoter, and MPG is located on both methylated and unmethylated promoters; and (iii) after completion of the repair, the MBD1-MPG complex is restored on the methylated promoter. Mobility-shift and structural analyses show that the MBD of MBD1 binds a methyl-CpG pair (mCpG x mCpG) but not the methyl-CpG pair containing a single 7-methylguanine (N) (mCpG x mCpN) that is known as one of the major lesions caused by MMS. We further demonstrate that knockdown of MBD1 by specific small interfering RNAs significantly increases cell sensitivity to MMS. These data suggest that MBD1 cooperates with MPG for transcriptional repression and DNA repair. We hypothesize that MBD1 functions as a reservoir for MPG and senses the base damage in chromatin
Figures
Fig. 1.
Interaction of MBD1 with MPG. (A) Structure of MBD1 and MPG. MBD1 (isoform v3) contains the MBD, nuclear localization signal (NLS), cysteine-rich CXXC domains, and TRD (9, 37). TRD (amino acids 473–536) was used in a yeast two-hybrid screen as a bait. MPG has an enzymatically active site at glutamic acid (E) 125. In full-length and deletion mutants of MPG, the presence and absence of MBD1 interaction in Fig. 2 A are indicated by plus and minus, respectively. (B) Association between endogenous MBD1 and MPG in HeLa cells. (C) Direct binding of the TRD of MBD1 to MPG in vitro. Bacterially expressed (His)6-TRD of MBD1 and GST-MPG were used for nickel-NTA resin affinity chromatography (38). The arrowhead indicates the full-length MPG fused to GST.
Fig. 2.
Regions of MPG for interacting with MBD1. (A) Association of MPG with the TRD of MBD1. GST-fused TRD of MBD1 or GST was immobilized on glutathione agarose beads and incubated with the lysates from HeLa cells expressing full-length or deletion mutants of MPG. (B) Localization of MPG and MBD1 in the nucleus. The full-length or deletion mutants of FLAG-MPG were coexpressed with DsRed-fused MBD1.
Fig. 3.
Cooperation of MBD1 and MPG for transcriptional repression. GAL4-MPG and FLAG-MBD1 (A), GAL4-TRD of MBD1 and FLAG-MPG (B), and GAL4-TRD of MBD1 and FLAG-MPG mutants (C) were expressed in HeLa cells to examine their effects on a luciferase reporter that contains five GAL4-binding elements just upstream of human SNRPN or the p16 gene promoter. The cells were transfected with plasmids (1 μg) to express FLAG-tagged proteins, together with indicated amounts of plasmids for GAL4-fused proteins. FLAG-mock was used as a control. The luciferase activities from insertless GAL4-mock were normalized to 100. Values are given as means and standard deviations of results from more than three independent experiments.
Fig. 4.
Molecular dynamics of MBD1 and MPG under MMS-induced DNA damage. (A) MMS-dependent growth inhibition of NCI-H1299 and SBC-5 cells. Cell numbers were examined after MMS treatment for 1 h and after incubation for 3 days. (B) Expression of MBD1, MPG, and β-tubulin in untreated and MMS-treated cells. At 48 h after transfection, both cell lines expressing HA-MBD1 and FLAG-MPG were treated with 1 mM MMS for 1 h. Western blot analysis was done with anti-MBD1, anti-FLAG, and anti-β-tubulin antibodies. (C) Association of MBD1 and MPG with chromosomal p16 gene promoters under MMS treatment. The p16 gene promoter is highly methylated in NCI-H1299 cells and unmethylated in SBC-5 cells (36, 37). For chromatin immunoprecipitation, the cells expressing HA-MBD1 and FLAG-MPG were treated with 1 mM MMS for 1 h and incubated for the indicated times. The coprecipitated DNAs with the indicated antibodies were PCR-amplified by using a set of primers for p16 promoter sequences. Genomic DNAs in the input cell lysates before the immunoprecipitation were used as a control. All data are from more than three independent experiments.
Fig. 5.
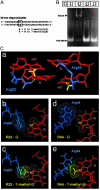
Abrogation of methylated DNA binding of MBD by the presence of 7-mG in a methyl–CpG pair. (A) Construction of 30 base-paired oligodeoxynucleotides containing 5-mC and/or 7-mG. The unique dinucleotides, CG × CG, mCG × mCG, mCG × mCmG, and CG × CmG, were introduced into position (XG × XY). mG or G was incorporated into position Y of the synthesized strand. (B) Electrophoretic mobility-shift assay by using the oligodeoxynucleotides and MBD from MBD1. (C) Structural model of the interaction between MBD and methylated DNA. A proposed interaction between the methyl–CpG site and two arginine side chains at the interface, on the basis of NMR structure determination, is shown in A. The interactions between the guanine bases and the arginine side chains (b and d) can be perturbed by alkylations at N7 positions of the guanine bases (c and e). Arginine 22 and 44, blue; DNA, red; hydrogen bond between arginine and guanine, white; methyl groups in methyl–CpG sequence, yellow.
Fig. 6.
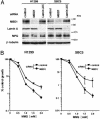
MBD1 knockdown increases cell sensitivity to MMS. (A) Effectiveness of siRNA knockdown of MBD1. The cells were transfected with siRNAs targeting mRNAs encoding MBD1 and lamin A/C. MPG and β-tubulin are also detected. (B) Effect of MBD1 knockdown on cell growth inhibition by MMS. Growth inhibition assay was performed as in Fig. 4_A_. GL3 siRNA was used as a control (41). Values are given as means and standard deviations from five independent experiments.
Similar articles
- Methyl-CpG binding domain proteins and their involvement in the regulation of the MAGE-A1, MAGE-A2, MAGE-A3, and MAGE-A12 gene promoters.
Wischnewski F, Friese O, Pantel K, Schwarzenbach H. Wischnewski F, et al. Mol Cancer Res. 2007 Jul;5(7):749-59. doi: 10.1158/1541-7786.MCR-06-0364. Mol Cancer Res. 2007. PMID: 17634428 - Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms.
Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Fujita N, et al. Mol Cell Biol. 1999 Sep;19(9):6415-26. doi: 10.1128/MCB.19.9.6415. Mol Cell Biol. 1999. PMID: 10454587 Free PMC article. - A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer.
Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. Lopez-Serra L, et al. Cancer Res. 2006 Sep 1;66(17):8342-6. doi: 10.1158/0008-5472.CAN-06-1932. Cancer Res. 2006. PMID: 16951140 - Regulation of transcription and chromatin by methyl-CpG binding protein MBD1.
Nakao M, Matsui S, Yamamoto S, Okumura K, Shirakawa M, Fujita N. Nakao M, et al. Brain Dev. 2001 Dec;23 Suppl 1:S174-6. doi: 10.1016/s0387-7604(01)00348-5. Brain Dev. 2001. PMID: 11738867 Review. - Preventing transcriptional gene silencing by active DNA demethylation.
Kapoor A, Agius F, Zhu JK. Kapoor A, et al. FEBS Lett. 2005 Oct 31;579(26):5889-98. doi: 10.1016/j.febslet.2005.08.039. Epub 2005 Aug 31. FEBS Lett. 2005. PMID: 16162337 Review.
Cited by
- Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2).
Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Valinluck V, et al. Nucleic Acids Res. 2004 Aug 9;32(14):4100-8. doi: 10.1093/nar/gkh739. Print 2004. Nucleic Acids Res. 2004. PMID: 15302911 Free PMC article. - Association between forkhead box P3 expression level and gender of Iranian juvenile idiopathic arthritis patients.
Ghavidel AA, Farivar S, Aghamiri S, Shiari R. Ghavidel AA, et al. Reumatologia. 2022;60(1):26-34. doi: 10.5114/reum.2022.114228. Epub 2022 Feb 28. Reumatologia. 2022. PMID: 35645413 Free PMC article. - Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase.
Baldwin MR, O'Brien PJ. Baldwin MR, et al. Biochemistry. 2009 Jun 30;48(25):6022-33. doi: 10.1021/bi900517y. Biochemistry. 2009. PMID: 19449863 Free PMC article. - Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials.
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Cheng Y, et al. Signal Transduct Target Ther. 2019 Dec 17;4:62. doi: 10.1038/s41392-019-0095-0. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 31871779 Free PMC article. Review. - A DNA repair-independent role for alkyladenine DNA glycosylase in alkylation-induced unfolded protein response.
Milano L, Charlier CF, Andreguetti R, Cox T, Healing E, Thomé MP, Elliott RM, Samson LD, Masson JY, Lenz G, Henriques JAP, Nohturfft A, Meira LB. Milano L, et al. Proc Natl Acad Sci U S A. 2022 Mar 1;119(9):e2111404119. doi: 10.1073/pnas.2111404119. Proc Natl Acad Sci U S A. 2022. PMID: 35197283 Free PMC article.
References
- Bird, A. (2002) Genes Dev. 16 6–21. - PubMed
- Panning, B. & Jaenisch, R. (1998) Cell 93 305–308. - PubMed
- Okano, M., Bell, D. W., Haber, D. A. & Li, E. (1999) Cell 99 247–257. - PubMed
- Ben-Porath, I. & Cedar, H. (2000) Curr. Opin. Genet. Dev. 10 550–554. - PubMed
- Baylin, S. B., Herman, J. G., Graff, J. R., Vertino, P. M. & Issa, J. P. (1998) Adv. Cancer Res. 72 141–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials