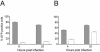Expression of the zinc-finger antiviral protein inhibits alphavirus replication - PubMed (original) (raw)
Expression of the zinc-finger antiviral protein inhibits alphavirus replication
Matthew J Bick et al. J Virol. 2003 Nov.
Abstract
The rat zinc-finger antiviral protein (ZAP) was recently identified as a host protein conferring resistance to retroviral infection. We analyzed ZAP's ability to inhibit viruses from other families and found that ZAP potently inhibits the replication of multiple members of the Alphavirus genus within the Togaviridae, including Sindbis virus, Semliki Forest virus, Ross River virus, and Venezuelan equine encephalitis virus. However, expression of ZAP did not induce a broad-spectrum antiviral state as some viruses, including vesicular stomatitis virus, poliovirus, yellow fever virus, and herpes simplex virus type 1, replicated to normal levels in ZAP-expressing cells. We determined that ZAP expression inhibits Sindbis virus replication after virus penetration and entry, but before the amplification of newly synthesized plus strand genomic RNA. Using a temperature-sensitive Sindbis virus mutant expressing luciferase, we further showed that translation of incoming viral RNA is blocked by ZAP expression. Elucidation of the antiviral mechanism by which ZAP inhibits Sindbis virus translation may lead to the development of agents with broad activity against alphaviruses.
Figures
FIG. 1.
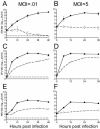
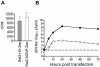
ZAP inhibits multiple members of the Alphavirus genus. Rat2-HA-Zeo cells expressing vector alone (filled circles) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open circles) were infected with SIN (A and B), SFV (C and D), or RRV (E and F) at MOIs of 0.01 and 5, as indicated. SIN MOIs were calculated based on stock titers determined on BHK-J cells, while SFV and RRV MOIs were based on stock titers determined on Rat2-HA-Zeo cells. At the indicated times after infection, medium was harvested and virus growth was determined by titration in duplicate on permissive cells. A separate well was utilized for each time point, and each experiment was done in duplicate. Dashed lines (A, C, and E), plaque assay detection limit. Data are mean log titers ± standard errors of the means; error bars for some points are obscured by the symbol.
FIG. 2.
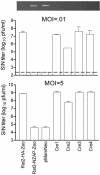
NZAP-Zeo expression is necessary for the resistance of Rat2-NZAP-Zeo cells to SIN infection. Rat2-HA-Zeo, Rat2-NZAP-Zeo, Rat2-NZAP-Zeo cells stably transfected with pMAMNeo (pMamNeo) and clones of Rat2-NZAP-Zeo cells stably transfected with pMAMNeo in combination with pMC-Cre (Cre1, -2, -3, and -4) were infected with SIN at MOIs of 0.01 and 5 as indicated, and the virus produced after 24 h was determined by titration in duplicate. Dashed line, plaque assay detection limit. Each cell line was tested in duplicate; data are mean log titers ± standard errors of the means. The retroviral insert present in each of the tested cell populations was amplified by PCR, and the ethidium-stained gel is shown at the top.
FIG. 3.
ZAP inhibits replication of a GFP-expressing VEE replicon. Rat2-HA-Zeo cells expressing vector alone (shaded bars) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open bars) were seeded in 12-well plates and infected with packaged SIN (A) or VEE (B) replicons expressing GFP. The percentage of cells expressing GFP was determined by flow cytometry at the indicated times after infection. Each bar represents the mean of results for three independent wells, with error bars indicating the standard deviations. The unpaired two-tailed t test indicates a significant difference between the mean values obtained after VEE infection of Rat2-HA-Zeo cells and those obtained after infection of Rat2-NZAP-Zeo cells (P < 0.0001 at 4 h, P = 0.0002 at 20 h). The results shown are similar to those from one other independent experiment.
FIG. 4.
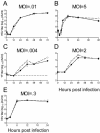
HSV-1, YF, and VSV are not inhibited by ZAP. Rat2-HA-Zeo cells expressing vector alone (filled circles) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open circles) were infected at the indicated MOIs with HSV-1 (A and B), YF (C and D), and VSV (E), and virus growth was determined by titration in duplicate on permissive cells. MOIs were calculated based on stock titers determined on Vero (HSV-1), BHK-J (YF), or Rat2-HA-Zeo (VSV) cells. A separate well was utilized for each time point, and the experiment was done in duplicate. Dashed line (C), plaque assay detection limit. Data are mean log titers ± standard errors of the means; error bars for some points are obscured by the symbol.
FIG. 5.
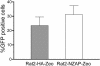
Replication of a poliovirus replicon is not inhibited by ZAP. Rat2-HA-Zeo cells expressing vector alone (shaded bars) and Rat2-NZAP-Zeo cells expressing the amino-terminal portion of ZAP fused to the product of the zeocin resistance gene (open bars) were seeded in six-well plates and transfected with 3.5 μg of PolioRep/GFP RNA encoding a GFP-expressing poliovirus replicon per well. The percentage of cells expressing GFP was determined by flow cytometry 8 h after transfection. Each bar represents the mean of results for three independent wells, with error bars indicating the standard deviations. Similar results were obtained in two independent experiments.
FIG. 6.
ZAP affects SIN replication after binding, penetration, and uncoating. (A) SIN binding. Rat2-HA-Zeo (shaded bar) and Rat2-NZAP-Zeo (open bar) cells were incubated in 12-well plates as described in Materials and Methods with 10,000 cpm of radioactive SIN (∼100 PFU/cell) for 1 h at 4°C to allow binding without penetration of SIN. Cells were washed extensively to remove unbound virus, and, after lysis in sodium dodecyl sulfate, the amount of bound virus was determined by counting an aliquot of the lysate. The experiment was done in triplicate and is representative of three independent experiments. Data are means, with error bars indicating the standard deviations. (B) SIN RNA transfection. SIN RNA was introduced via cationic lipid transfection into Rat2-HA-Zeo cells expressing vector alone (filled circles) and Rat2-NZAP-Zeo cells (open circles). At the indicated times after transfection, an aliquot of the medium was harvested, and virus growth was determined by titration in duplicate on permissive cells. Dashed line, plaque assay detection limit. The experiment was done in duplicate and is representative of two independent experiments. Data are mean log titers ± standard errors of the means; error bars for some points are obscured by the symbol.
FIG. 7.
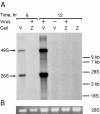
ZAP blocks the production of new plus strand genomic RNA. (A) Rat2-HA-Zeo (V) and Rat2-NZAP-Zeo (Z) cells were mock-infected (−) or infected (+) with SIN (MOI = 5, as titered on BHK-J cells) in the presence of actinomycin D (1 μg/ml) as described in Materials and Methods. After infection, [3H]uridine was added, and RNA was harvested at the indicated times after infection. Total RNA (5 μg) was size separated by denaturing gel electrophoresis, and, after ethidium bromide staining, the gel was treated for fluorography and exposed to film for 28 h at −80°C. On prolonged exposure (1 week) a faint 49S signal, likely due to a small percentage of permissive cells in the culture, can be seen in infected Rat2-NZAP-Zeo cells harvested at 12 h. Migration of RNA size markers is shown on the right. Arrows, locations of the SIN genomic (49S) and subgenomic (26S) RNAs. (B) Ethidium bromide staining of ribosomal 28S RNA prior to fluorography indicates equal loading of RNA on the gel. The results are representative of two independent experiments.
FIG. 8.
ZAP blocks translation of incoming SIN RNA. (A) Schematic representation of Toto1101 virus and luciferase-expressing variants. Solid lines, UTRs. Open and shaded boxes, locations of the nonstructural- and structural-protein-encoding regions, respectively. The approximate locations of the nonstructural proteins nsP1, nsP2, nsP3, and nsP4 and structural protein capsid (C), glycoproteins (PE and E1), and the 6-kDa protein are shown, as is the location of the cap and poly(A) tail (An). _Spe_I indicates the restriction site used for cloning the luciferase (Luc) gene into pToto1101. Hatched bars within nsP3, firefly luciferase-encoding region. The 36-nt deletion resulting in a defective RNA-dependent RNA polymerase is indicated (Δ_Kpn_I), as are the glycine (G)-to-glutamic acid (E) mutations of ts6 and ts110. (B) Rat2-HA-Zeo control cells (shaded bars) and Rat2-NZAP-Zeo cells (open bars) were infected with firefly luciferase-expressing SIN temperature-sensitive mutant Toto1101/Luc:ts6 (MOI = 0.006). Binding was carried out at 4°C. After being washed, the cells were incubated at the nonpermissive temperature (40°C). At the indicated times after infection, the cells were lysed and luciferase activity was measured with the luciferase assay system (Promega). (C and D) Rat2-HA-Zeo control cells (shaded bars) and Rat2-NZAP-Zeo cells (open bars) were transfected for 30 min with a mixture of 0.9 μg of capped Toto1101/Luc/Pol− RNA and 0.1 μg of capped control RNA encoding Renilla luciferase. Cells were lysed at the indicated times, and Renilla (C) and firefly (D) luciferase activities were measured with the Dual-Luciferase reporter assay system (Promega). Renilla luciferase activity indicates translation of the control RNA (C), while firefly luciferase activity indicates translation of SIN RNA (D). For panels B to D the data are representative of two independent experiments. Bars represent the means of triplicate samples ± standard deviations. RLU, relative light units.
Similar articles
- The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses.
MacDonald MR, Machlin ES, Albin OR, Levy DE. MacDonald MR, et al. J Virol. 2007 Dec;81(24):13509-18. doi: 10.1128/JVI.00402-07. Epub 2007 Oct 10. J Virol. 2007. PMID: 17928353 Free PMC article. - Sindbis Virus Can Exploit a Host Antiviral Protein To Evade Immune Surveillance.
Wang X, Li MMH, Zhao J, Li S, MacDonald MR, Rice CM, Gao X, Gao G. Wang X, et al. J Virol. 2016 Oct 28;90(22):10247-10258. doi: 10.1128/JVI.01487-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581990 Free PMC article. - Alphavirus Evasion of Zinc Finger Antiviral Protein (ZAP) Correlates with CpG Suppression in a Specific Viral nsP2 Gene Sequence.
Nguyen LP, Aldana KS, Yang E, Yao Z, Li MMH. Nguyen LP, et al. Viruses. 2023 Mar 24;15(4):830. doi: 10.3390/v15040830. Viruses. 2023. PMID: 37112813 Free PMC article. - Versatility of the Zinc-Finger Antiviral Protein (ZAP) As a Modulator of Viral Infections.
Shao R, Visser I, Fros JJ, Yin X. Shao R, et al. Int J Biol Sci. 2024 Aug 26;20(12):4585-4600. doi: 10.7150/ijbs.98029. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309436 Free PMC article. Review. - A structural and functional perspective of alphavirus replication and assembly.
Jose J, Snyder JE, Kuhn RJ. Jose J, et al. Future Microbiol. 2009 Sep;4(7):837-56. doi: 10.2217/fmb.09.59. Future Microbiol. 2009. PMID: 19722838 Free PMC article. Review.
Cited by
- Neurotropic arboviruses induce interferon regulatory factor 3-mediated neuronal responses that are cytoprotective, interferon independent, and inhibited by Western equine encephalitis virus capsid.
Peltier DC, Lazear HM, Farmer JR, Diamond MS, Miller DJ. Peltier DC, et al. J Virol. 2013 Feb;87(3):1821-33. doi: 10.1128/JVI.02858-12. Epub 2012 Nov 28. J Virol. 2013. PMID: 23192868 Free PMC article. - Antiviral Activity of Zinc Finger Antiviral Protein (ZAP) in Different Virus Families.
de Andrade KQ, Cirne-Santos CC. de Andrade KQ, et al. Pathogens. 2023 Dec 17;12(12):1461. doi: 10.3390/pathogens12121461. Pathogens. 2023. PMID: 38133344 Free PMC article. Review. - The Antiviral Activities of Poly-ADP-Ribose Polymerases.
Malgras M, Garcia M, Jousselin C, Bodet C, Lévêque N. Malgras M, et al. Viruses. 2021 Mar 30;13(4):582. doi: 10.3390/v13040582. Viruses. 2021. PMID: 33808354 Free PMC article. Review. - LINE-1 Retroelements Get ZAPped!
McLaughlin RN Jr, Malik HS. McLaughlin RN Jr, et al. PLoS Genet. 2015 Jul 16;11(7):e1005364. doi: 10.1371/journal.pgen.1005364. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26182081 Free PMC article. No abstract available. - Gamma interferon-dependent, noncytolytic clearance of sindbis virus infection from neurons in vitro.
Burdeinick-Kerr R, Griffin DE. Burdeinick-Kerr R, et al. J Virol. 2005 May;79(9):5374-85. doi: 10.1128/JVI.79.9.5374-5385.2005. J Virol. 2005. PMID: 15827152 Free PMC article.
References
- Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. - PubMed
- Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891-1899. - PubMed
- Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703-1706. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases