Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit - PubMed (original) (raw)
Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
T Martin Schmeing et al. RNA. 2003 Nov.
Abstract
During translation, tRNAs cycle through three binding sites on the ribosome: the A, the P, and the E sites. We have determined the structures of complexes between the Haloarcula marismortui large ribosomal subunit and two different E site substrates: a deacylated tRNA acceptor stem minihelix and a CCA-acceptor end. Both of these tRNA mimics contain analogs of adenosine 76, the component responsible for a large proportion of E site binding affinity. They bind in the center of the loop-extension of protein L44e, and make specific contacts with both L44e and 23S rRNA including bases that are conserved in all three kingdoms of life. These contacts are consistent with the footprinting, protection, and cross-linking data that have identified the E site biochemically. These structures explain the specificity of the E site for deacylated tRNAs, as it is too small to accommodate any relevant aminoacyl-tRNA. The orientation of the minihelix suggests that it may mimic the P/E hybrid state. It appears that the E site on the 50S subunit was formed by only RNA in the last common ancestor of the three kingdoms, since the proteins at the E sites of H. marismortui and Deinucoccus radiodurans large subunits are not homologous.
Figures
FIGURE 1.
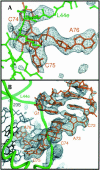
Experimental electron density of E site ligands bound to the H. marismortui 50S ribosomal subunit. (A) Fo(complex)–Fo(parent) electron density map of CCA bound to the 50S subunit, calculated with experimental phases, contoured at 4 σ. Electron density is visible for the portion of ribosomal protein L44e that adopts an altered conformation upon tRNA binding, near the top of the frame. (B) Fo(complex)–Fo(parent) electron density map of the minihelix bound to the 50S subunit, using experimental phases, contoured at 3 σ. Electron density gets weaker further from the CCA end because of increasing disorder. Density for the mobile portion of L44e is at the bottom of the frame.
FIGURE 2.
Overview of the tRNA binding sites on the large subunit. CCA oligomers are shown bound at the A site (red) and P site (blue), and the minihelix is shown bound at the E site (orange). 23S rRNA is colored gray and ribosomal proteins are green. Proteins are shown in ribbon format, and RNA all atom, with backbones in darker and bases in lighter colors. The independently solved complex of the L1 arm and L1 protein (Nikulin et al. 2003) was docked into experimental electron density maps of the H. marismortui 50S subunit calculated at 9 Å resolution (Ban et al. 1998).
FIGURE 3.
L44e interacts with the E-site-bound minihelix. (A) The final 2 nt of the minihelix insert through the loop extension of L44e. (B) L44e undergoes a conformational change upon E site tRNA binding, from the conformation seen in yellow to that in green. Proline 56 is shown in both panels A and B to show relative orientation.
FIGURE 4.
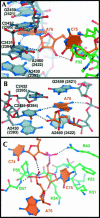
Specific interactions of E site binding. (A) The E site ligands interact with both rRNA (gray) and L44e (green). (B) A76 stacks in between 23S rRNA nucleotides G2459 (2421) and A2460 (2422), and hydrogen bonds with universally conserved C2431 (2394). (C) C75 interacts with L44e through hydrogen bonding to Gly 57 and Arg 40, and hydrophobic packing with Lys 54, Lys 51, and Phe 52.
FIGURE 5.
Comparison with tRNA bound to the E site in the 70S structure. (A) The minihelix (orange) is in a position similar to the tRNA (green) bound to the 70S ribosome E site tRNA. Corresponding phosphorous atoms of the H. marismortui 50S subunit and T. thermophilus 70S ribosome were superimposed to allow the comparison. (B) A yeast tRNAphe (red) superimposed onto the duplex portion of the minihelix (orange) has its anticodon stem–loop closer to the anticodon stem loop of a tRNA bound in the 30S P site (blue) than a tRNA bound in the 30S E site (green).
FIGURE 6.
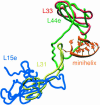
Differences in eubacterial and archeal E site ribosomal proteins. The globular portions of H. marismortui ribosomal proteins L44e (green) and L15e (blue) are replaced by L33 (red) and L31 (yellow), respectively, in the D. radiodurans ribosome (Harms et al. 2001). However, the loop portion of L44e that interacts with the 3′ terminus of the E site-bound minihelix (orange) overlaps with an extension of L31.
Similar articles
- Transit of tRNA through the Escherichia coli ribosome: cross-linking of the 3' end of tRNA to ribosomal proteins at the P and E sites.
Kirillov SV, Wower J, Hixson SS, Zimmermann RA. Kirillov SV, et al. FEBS Lett. 2002 Mar 6;514(1):60-6. doi: 10.1016/s0014-5793(02)02302-5. FEBS Lett. 2002. PMID: 11904182 - An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA.
Schmeing TM, Huang KS, Strobel SA, Steitz TA. Schmeing TM, et al. Nature. 2005 Nov 24;438(7067):520-4. doi: 10.1038/nature04152. Nature. 2005. PMID: 16306996 - The structural basis of ribosome activity in peptide bond synthesis.
Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Nissen P, et al. Science. 2000 Aug 11;289(5481):920-30. doi: 10.1126/science.289.5481.920. Science. 2000. PMID: 10937990 - RNA, the first macromolecular catalyst: the ribosome is a ribozyme.
Steitz TA, Moore PB. Steitz TA, et al. Trends Biochem Sci. 2003 Aug;28(8):411-8. doi: 10.1016/S0968-0004(03)00169-5. Trends Biochem Sci. 2003. PMID: 12932729 Review. - The 30S ribosomal P site: a function of 16S rRNA.
Noller HF, Hoang L, Fredrick K. Noller HF, et al. FEBS Lett. 2005 Feb 7;579(4):855-8. doi: 10.1016/j.febslet.2004.11.026. FEBS Lett. 2005. PMID: 15680962 Review.
Cited by
- Contribution of ribosomal residues to P-site tRNA binding.
Shoji S, Abdi NM, Bundschuh R, Fredrick K. Shoji S, et al. Nucleic Acids Res. 2009 Jul;37(12):4033-42. doi: 10.1093/nar/gkp296. Epub 2009 May 5. Nucleic Acids Res. 2009. PMID: 19417061 Free PMC article. - Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab.
Webb KJ, Laganowsky A, Whitelegge JP, Clarke SG. Webb KJ, et al. J Biol Chem. 2008 Dec 19;283(51):35561-8. doi: 10.1074/jbc.M806006200. Epub 2008 Oct 28. J Biol Chem. 2008. PMID: 18957409 Free PMC article. - An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination.
Amort M, Wotzel B, Bakowska-Zywicka K, Erlacher MD, Micura R, Polacek N. Amort M, et al. Nucleic Acids Res. 2007;35(15):5130-40. doi: 10.1093/nar/gkm539. Epub 2007 Jul 26. Nucleic Acids Res. 2007. PMID: 17660192 Free PMC article. - The CCA-end of P-tRNA Contacts Both the Human RPL36AL and the A-site Bound Translation Termination Factor eRF1 at the Peptidyl Transferase Center of the Human 80S Ribosome.
Hountondji C, Bulygin K, Créchet JB, Woisard A, Tuffery P, Nakayama J, Frolova L, Nierhaus KH, Karpova G, Baouz S. Hountondji C, et al. Open Biochem J. 2014 Jul 11;8:52-67. doi: 10.2174/1874091X01408010052. eCollection 2014. Open Biochem J. 2014. PMID: 25191528 Free PMC article. - Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations.
Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, Korostelev AA. Svidritskiy E, et al. Structure. 2014 Aug 5;22(8):1210-1218. doi: 10.1016/j.str.2014.06.003. Epub 2014 Jul 17. Structure. 2014. PMID: 25043550 Free PMC article.
References
- Agrawal, R.K., Penczek, P., Grassucci, R.A., Li, Y., Leith, A., Nierhaus, K.H., and Frank, J. 1996. Direct visualization of A-, P-, and E-site transfer RNAs in the Escherichia coli ribosome. Science 271: 1000–1002. - PubMed
- Ban, N., Freeborn, B., Nissen, P., Penczek, P., Grassucci, R.A., Sweet, R., Frank, J., Moore, P.B., and Steitz, T.A. 1998. A 9 Å resolution X-ray crystallographic map of the large ribosomal subunit. Cell 93: 1105–1115. - PubMed
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. - PubMed
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources