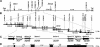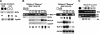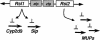Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression - PubMed (original) (raw)
. 2003 Nov 1;17(21):2664-74.
doi: 10.1101/gad.1135703. Epub 2003 Oct 16.
Affiliations
- PMID: 14563677
- PMCID: PMC280616
- DOI: 10.1101/gad.1135703
Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression
Christopher J Krebs et al. Genes Dev. 2003.
Abstract
Sexually dimorphic expression of a broad array of liver proteins involved in reproduction and xenobiotic metabolism is induced at puberty by sex-specific growth hormone patterns. An additional control of sex-dependent gene expression is conferred by Regulator of sex-limitation (Rsl) alleles. In variant rsl mice, females inappropriately express the male Sex-limited protein, Slp. We recently showed that a panel of male-specific liver genes is repressed by Rsl, accentuating sex differences in a hormone-independent manner. Here we map rsl to a region on Chromosome 13 comprised exclusively of KRAB (Kruppel-associated box) zinc-finger protein (ZFP) genes. Among eight Rsl candidate (Rslcan) genes within the critical genetic interval, the recent duplicates Rslcan-4 and Rslcan-9 both harbor mutations in rsl mice (partial deletion and splice-site inactivation, respectively). Transgenesis with bacterial artificial chromosome (BAC) clones encompassing Rslcan-4 restores male-specific MUP (major urinary protein) expression to rsl mice, whereas a BAC containing Rslcan-9 rescues sex-specific expression of Slp and cytochrome P450 Cyp2d9. Thus, the Rslcan-4 and Rslcan-9 paralogs partitioned regulation of their target genes during evolution. This demonstrates the first biological role for a set of KRAB zinc-finger repressor proteins and reveals the molecular basis of a gene-silencing pathway critical for sexual dimorphism.
Figures
Figure 1.
Rsl variation revealed by differential effects on male-specific target genes. (A) Total liver RNA from congenic and inbred mice was assayed for Slp by RNase protection (RPA), using a probe that also detects Slp's homolog C4 (Cox and Robins 1988). Elevated Slp levels in B10.D2.PL, B10.D2.FM, and 129, relative to B10.D2, females are hallmarks of the rsl phenotype. rsl allele-specific differences are evident in lower Slp levels for rslFM than rslPL mice. Allelic variation also exists in target genes; for example, B6 mice do not express Slp. C4 is an internal control that is not sex-dependent. (B) Mouse urinary proteins (MUPs) were analyzed by Northern blot of liver RNAs using an oligonucleotide probe complementary to MUP III (Shahan et al. 1987), which is elevated in both sexes of rslPL mice (more obvious in females because of their normally low expression), relative to B10.D2 mice. B6 mice are Rsl based on dimorphism for MUP III. The filter was reprobed with GAPDH cDNA as a loading control. (C) Summary of phenotypes. Strains with strongly dimorphic expression of male-specific liver genes are considered wild type for Rsl; rsl strains are those in which females have lost regulation of sex-limitation of liver target genes.
Figure 2.
High-resolution genetic map of the Rsl locus. Using a PL/J × B10.D2 intercross, 1660 F2 mice were typed with polymorphic microsatellite markers (listed at left). D13Mit markers were from Research Genetics; D13Dmr markers were developed from the B6 BAC contig (see Fig. 3). Open boxes indicate homozygosity for the PL/J allele, filled boxes homozygosity for the B10.D2 allele, and diagonally half-filled boxes indicate heterozygosity. At the bottom is the number of recombinants that share the haplotype indicated. Scoring for liver Slp expression in directly informative females (those having a homozygous PL/J flanking marker) is shown on the line below (♀ Slp), indicated as positive (+) or negative (-). Within this group, the recombinant female with a breakpoint between D13Mit124 and D13Dmr1 defined the centromeric (top) side of the Rsl interval. For all other recombinants, testing of female backcross progeny is shown below the arrows as the proportion positive for Slp. The telomeric side of the Rsl interval, between D13Dmr8 and D13Dmr5, was defined by the single recombinant with 7 of 15 female progeny positive for Slp.
Figure 3.
Physical mapping of the Rsl locus. (A) A genetic map was derived by genotyping eight MIT microsatellite markers in 15 recombinant mice. The recombination frequencies are shown in centimorgans between markers. The previously mapped growth arrest specific gene 1 (gas1) and adenylate cyclase 2 (adcy2) are indicated. (B) Six BACs (solid lines, with address names below) form a contig spanning the Rsl locus. The 16 BAC-derived microsatellite markers and two polymorphic Rsl candidate genes (Rslcan-8 and Rslcan-6) used in mapping are indicated above the contig. Black boxes indicate Rsl candidate KRAB zinc-finger genes, numbered in the order of their discovery. Mzf22 is the only previously reported gene in this region, along with Arbp, a processed pseudogene. (C) Schematic of eight candidate genes within the critical genetic interval. Shade or pattern indicates their pairwise similarity. Rslcan-17 is most similar to Rslcan-14, Rslcan-9 to Rslcan-4, Rslcan-10 to Rslcan-13, and Rslcan-7 to Rslcan-6. The Human Genome Nomenclature Committee has assigned names to these genes (Zfps455-460 and Rsl1 and Rsl2) in accord with functional identification of the latter two genes in this report. Arrowheads indicate the direction of transcription. (D) Genomic architecture of a prototypical KRAB zinc-finger gene from the Rsl locus. Black boxes represent the coding regions within the exons, with approximate distances (in kilobases) between exons given. ATG and TGA indicate start and stop codons, respectively. Exon 1 typically encodes a short (∼2-12-amino acid) N-terminal leader, prior to the highly conserved KRAB A domain, which starts immediately within exon 2. KRAB A and B domains encompass repressor activity, tethered to DNA via a variable number of zinc fingers located in the fourth and last exon.
Figure 4.
Deletion of Rslcan-4 sequences by recombination with Rslcan-13 in rsl mice. (A) The common structure of Rslcan genes is shown, with PCR primers flanking the zinc fingers and a _Pst_I site unique to Rslcan-9 in parentheses. Below are the genomic _Pst_I fragments revealed by a probe that cross-hybridizes with Rslcan-4, Rslcan-9, and the adjacent Rslcan-13. (B) Map of overlapping BACs from B6 and 129 libraries (BAC addresses are on the left). Numbered black boxes indicate Rslcan genes determined by PCR. An open box and arrow indicate the Rslcan-4/Rslcan-13 hybrid discovered by sequencing. (C) PCR products from zinc-finger regions of Rslcan-4 and Rslcan-9 were digested with _Pst_I; Rslcan-4 remains undigested (∼1.4 kb), while Rslcan-9 is cleaved into bands of 942 and 450 bp. PCR products were analyzed from B10.D2, B10.D2.PL, and 129 mouse DNAs and BACs 349-F-18 and 163-B-15. The absence of Rslcan-4 bands from B10.D2.PL and 129 mice suggests the gene is deleted in rsl strains. (D) Southern blot of BAC (left) and mouse genomic (right) DNAs hybridized with an Rslcan-4 zinc-finger probe shows two large bands in BAC 163-B-15 and in genomic DNAs from B10.D2 and B6 mice, representing Rslcan-4 and Rslcan-13 fragments. A single intermediate band in BAC 79-I-2 and genomic DNAs from B10.D2.PL and 129 mice corroborates fusion of the genes to an Rslcan-4/Rslcan-13 hybrid in rsl mice. (E) In aligned sequences of Rslcan-4 and Rslcan-13 with B10.D2.PL genomic DNA, shading indicates regions of alternating identity between B10.D2.PL and Rslcan-4 or Rslcan-13. The start codon (open box) and amino acids encoded in exon 1 of Rslcan-4 are shown. Filled circles highlight single-base differences; black boxes are bases unique to B10.D2.PL. The alternating regions of identity extend for an additional 10 kb, resolving into complete identity with Rslcan-13 near the 5′-end of the zinc-finger domain in exon 4. Genomic sequence from B10.D2.PL DNA is identical to 129 in this region, even for the B10.D2.PL-specific bases.
Figure 5.
Identification of a splice-site mutation in Rslcan-9. (A) The SS1 splice donor sequence of the first intron of Rslcan-9 has a T at position +5 (IVS + 5) in rsl strains B10.D2.PL, B10.D2.FM, and 129, but a G in the consensus splice sequence and in Rsl mice (B10.D2, B6). (B) RT-PCR analysis of Rslcan-9 liver RNAs. Diagrammed in C is the Rslcan-9 gene with splice donor sites (SS1 and SS2) identified in exon 1 and arrows indicating primers used for RT-PCR. Both transcripts are present in Rsl strains. The absence of the SS1 transcript in rsl strains is apparently the consequence of the IVS + 5 mutation in SS1. (C) The 5′-ends of SS1 and SS2 transcripts are diagrammed. A hatched box represents a small open reading frame in the SS2 transcript that is out of frame with the KRAB-encoding exon and would likely interfere with translation from the second AUG. Both the first and second AUG codons are in frame in the SS1 transcript, suggesting this is the more translationally competent of the two.
Figure 6.
BAC transgenic rescue of the rsl phenotype. B6-derived BAC clones 163-B-15 and 45-N-22 were independently injected into oocytes, and transgenic mice were bred for homozygosity for PL/J markers flanking rsl. Transgenic mice (black dot in white circle) were identified by PCR of tail DNA for BAC sequences. To assess Rsl function, mouse urine was assayed for MUP protein by SDS-PAGE, liver mRNAs for MUP I/II and cyp2d9 by Northern blot, and Slp by RPA. GAPDH and cyp2a4 (female-prominent) were loading controls for Northern blots. C4 mRNA was an internal control for RPAs (data not shown). (A) Expression of MUP protein and Slp and cyp2d9 mRNA in inbred Rsl and rslPL strains, for comparison. Expression of all target genes is elevated in rsl mice, most visibly in females. (B, left) For BAC 163-B-15 (containing B6-derived Rslcan-4), transgenic mice show reduced MUP protein in urine and MUP I/II mRNA in liver, but still have high Slp levels, relative to their nontransgenic siblings. This effect is more pronounced in females (e.g., cf. the two transgenic and the nontransgenic females). These results indicate that Rslcan-4 can restore dimorphic expression of MUP, but not Slp (lower panel) or cyp2d9 (data not shown). (Right) In contrast, mice transgenic for BAC 45-N-22 (containing B6-derived Rslcan-9) show reduced expression of Slp and cyp2d9, but expression of MUP I/II remains elevated (again, cf. the two females in the rightmost lanes). (C) Mice from two additional independent BAC 45-N-22 transgenic lines were assayed for liver expression of Rslcan-9 by RT-PCR, as in Figure 5. BAC DNA and Slp mRNA were assayed as above. For comparison, results from nontransgenic Rsl (filled circle) and rsl (open circle) females are at left. Representative sisters from one line (black dot in white circle) show expression of Rslcan-9 SS1 transcript and extinguished Slp expression. In contrast, sisters of another line (black dot in hatched circle) only express SS2 and Slp remains elevated.
Figure 7.
Rsl function is partitioned between two paralogous KRAB-ZFP genes. These findings indicate a division of labor between the recently duplicated Rsl1 (Rslcan-9) and Rsl2 (Rslcan-4), whereby one represses expression of MUP genes, and the other represses Slp and cyp2d9. This action sums to enforce sexually dimorphic expression of a set of genes that are hormonally induced in males at puberty. Biological functions of other ZFP genes in the region remain to be determined.
Similar articles
- Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2.
Krebs CJ, Larkins LK, Khan SM, Robins DM. Krebs CJ, et al. Genomics. 2005 Jun;85(6):752-61. doi: 10.1016/j.ygeno.2005.03.004. Epub 2005 Apr 15. Genomics. 2005. PMID: 15885501 - A pair of mouse KRAB zinc finger proteins modulates multiple indicators of female reproduction.
Krebs CJ, Robins DM. Krebs CJ, et al. Biol Reprod. 2010 Apr;82(4):662-8. doi: 10.1095/biolreprod.109.080846. Epub 2009 Dec 30. Biol Reprod. 2010. PMID: 20042539 Free PMC article. - Regulator of sex-limitation KRAB zinc finger proteins modulate sex-dependent and -independent liver metabolism.
Krebs CJ, Khan S, MacDonald JW, Sorenson M, Robins DM. Krebs CJ, et al. Physiol Genomics. 2009 Jun 10;38(1):16-28. doi: 10.1152/physiolgenomics.90391.2008. Epub 2009 Apr 7. Physiol Genomics. 2009. PMID: 19351907 Free PMC article. - The regulator of sex-limitation gene, rsl, enforces male-specific liver gene expression by negative regulation.
Tullis KM, Krebs CJ, Leung JY, Robins DM. Tullis KM, et al. Endocrinology. 2003 May;144(5):1854-60. doi: 10.1210/en.2002-0190. Endocrinology. 2003. PMID: 12697692 - A KRABsody for Embryo-Placental Development.
Trono D. Trono D. Dev Cell. 2017 Jun 19;41(6):578-580. doi: 10.1016/j.devcel.2017.06.005. Dev Cell. 2017. PMID: 28633014 Review.
Cited by
- Strain-Specific Epigenetic Regulation of Endogenous Retroviruses: The Role of _Trans_-Acting Modifiers.
Elmer JL, Ferguson-Smith AC. Elmer JL, et al. Viruses. 2020 Jul 27;12(8):810. doi: 10.3390/v12080810. Viruses. 2020. PMID: 32727076 Free PMC article. Review. - Gender-specific induction of cytochrome P450s in nonylphenol-treated FVB/NJ mice.
Hernandez JP, Chapman LM, Kretschmer XC, Baldwin WS. Hernandez JP, et al. Toxicol Appl Pharmacol. 2006 Oct 15;216(2):186-96. doi: 10.1016/j.taap.2006.05.014. Epub 2006 May 25. Toxicol Appl Pharmacol. 2006. PMID: 16828826 Free PMC article. - Three Sgp loci act independently as well as synergistically to elevate the expression of specific endogenous retroviruses implicated in murine lupus.
Ito K, Baudino L, Kihara M, Leroy V, Vyse TJ, Evans LH, Izui S. Ito K, et al. J Autoimmun. 2013 Jun;43:10-7. doi: 10.1016/j.jaut.2013.01.014. Epub 2013 Mar 7. J Autoimmun. 2013. PMID: 23465716 Free PMC article. - Role of STAT5a in regulation of sex-specific gene expression in female but not male mouse liver revealed by microarray analysis.
Clodfelter KH, Miles GD, Wauthier V, Holloway MG, Zhang X, Hodor P, Ray WJ, Waxman DJ. Clodfelter KH, et al. Physiol Genomics. 2007 Sep 19;31(1):63-74. doi: 10.1152/physiolgenomics.00055.2007. Epub 2007 May 29. Physiol Genomics. 2007. PMID: 17536022 Free PMC article. - Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes.
Van Keuren ML, Gavrilina GB, Filipiak WE, Zeidler MG, Saunders TL. Van Keuren ML, et al. Transgenic Res. 2009 Oct;18(5):769-85. doi: 10.1007/s11248-009-9271-2. Epub 2009 Apr 26. Transgenic Res. 2009. PMID: 19396621 Free PMC article.
References
- Ausubel F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1993. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York.
- Bacchini A., Gaetani, E., and Cavaggioni, A. 1992. Pheromone binding proteins of the mouse Mus musculus. Experientia 48: 419-421. - PubMed
- Brown L.J. and Shreffler, D.C. 1980. Female expression of the H-2 linked Sex-limited protein (Slp) due to non-H-2 genes. Immunogenetics 10: 19-29. - PubMed
- Burge C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases