A presenilin dimer at the core of the gamma-secretase enzyme: insights from parallel analysis of Notch 1 and APP proteolysis - PubMed (original) (raw)
. 2003 Oct 28;100(22):13075-80.
doi: 10.1073/pnas.1735338100. Epub 2003 Oct 17.
Ma Xenia G Ilagan, Anne L Brunkan, Silva Hecimovic, Yue-ming Li, Min Xu, Huw D Lewis, Meera T Saxena, Bart De Strooper, Archie Coonrod, Taisuke Tomita, Takeshi Iwatsubo, Chad L Moore, Alison Goate, Michael S Wolfe, Mark Shearman, Raphael Kopan
Affiliations
- PMID: 14566063
- PMCID: PMC240747
- DOI: 10.1073/pnas.1735338100
A presenilin dimer at the core of the gamma-secretase enzyme: insights from parallel analysis of Notch 1 and APP proteolysis
Eric H Schroeter et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Notch receptors and the amyloid precursor protein are type I membrane proteins that are proteolytically cleaved within their transmembrane domains by a presenilin (PS)-dependent gamma-secretase activity. In both proteins, two peptide bonds are hydrolyzed: one near the inner leaflet and the other in the middle of the transmembrane domain. Under saturating conditions the substrates compete with each other for proteolysis, but not for binding to PS. At least some Alzheimer's disease-causing PS mutations reside in proteins possessing low catalytic activity. We demonstrate (i) that differentially tagged PS molecules coimmunoprecipitate, and (ii) that PS N-terminal fragment dimers exist by using a photoaffinity probe based on a transition state analog gamma-secretase inhibitor. We propose that gamma-secretase contains a PS dimer in its catalytic core, that binding of substrate is at a site separate from the active site, and that substrate is cleaved at the interface of two PS molecules.
Figures
Fig. 1.
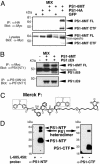
Notch and APP compete for a limiting activity but not for binding to PS1. (A) Saturation of γ-secretase was demonstrated by transfecting HEK293 cells with a mixture NΔE and cytoβ-gal DNA to a total of 5 μg (two independent experiments were done for each concentration shown). Cells were metabolically labeled overnight with 35S-Met, and cell extracts were immunoprecipitated with 9E10 antibody and separated by SDS/PAGE. The counts in each lane were determined by PhosphorImaging and plotted to show total Notch labeling (line graph) and percentage of NICD (NICD/NΔE + NICD; histograms) as a function of DNA concentration. (B and C) HEK293 cells were transfected with pCS2+C99 and Notch expressing plasmid or control plasmid. Aβ levels were assayed by ELISA. Percentage of NICD was determined by quantitative Western analysis and 35S labeling, which yielded similar results. Results show that APP competes with Notch for the same cleavage activity. NΔE and, more effectively, NΔE(V>L) compete for Aβ production although NLNR and NICD do not. (D) HEK293 cells were transiently transfected with GFP, NΔE, or NLNR, lysed in co-IP buffer, and immunoprecipitated with R22 antibody. PS1 NTF and CTF were detected via Western blotting with NT1 and 13A11 antibodies, respectively. (E) Notch proteins were detected in the same immunoprecipitate by using 9E10. NLNR and NΔE interact with PS1 as assayed by co-IP (equal protein input, data not shown). FC-NLNR, furin-cleaved NLNR; FL, full length.
Fig. 2.
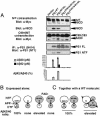
Co-IP and cross-linking analyses suggest dimerization of PS1 molecules within γ-secretase. (A) HEK293 cells were transfected with the indicated constructs, lysed, and immunoprecipitated with a polyclonal α-HA antibody (Covance). Immunoprecipitates and lysates were resolved by SDS/PAGE and immunoblotted with 9E10 antibodies. (B) PS1/PS2-deficient MEFs were transfected with PS1-6MT or PS1ΔE9 or both. They were then lysed, immunoprecipitated with 9E10 or 9N14 antibodies, and analyzed by immunostaining with NT1 antibody. As a control, IPs were also performed by using lysates from singly transfected cells mixed postlysis (labeled MIX). (C) Structure of Merck F (benzophenone photoreactive groups are indicated by boxes). (D) Covalent labeling of PS1 by photoreactive active site-directed γ-secretase inhibitor (Merck F). 3-[(3-Cholamidopropyl)dimethylammonio]-2-hydroxy-1-propane-sulfonate-solubilized HeLa cell membranes were photoactivated in the absence (-) or presence (+) of L-685,458 as a competitor. The sample was diluted with radioimmunoprecipitation assay buffer. Biotinylated proteins were captured with streptavidin-agarose and probed by SDS/PAGE/immunoblotting using α-PS1 antibodies. FL, full length.
Fig. 4.
A strategy for testing trans acting FAD activity. (A) WT and mutant forms of PS1 were transiently transfected into PS1/PS2-deficient MEFs and their associated proteolytic activities were assayed. Results show that mutating Asp-385 and Asp-257 in the FAD mutant PS1M146L abolishes NICD and AICD release, Aβ formation, and endoproteolysis. For Aβ measurements, average of independent triplicates from one representative experiment is shown (n > 10). (B) A schematic showing the expected Aβ42/Aβ40 ratio associated with different PS molecules when expressed alone (i.e., intrinsic activity). (C) Coexpression of the double mutant PS1D385A/M146L with PS1WT is predicted to distinguish between a monomeric (Left) or dimeric (Right) acting PS within the catalytic core of γ-secretase. If PS strictly acts as monomer the loss of Asp-385 should be epistatic to the intramolecular FAD mutation and the Aβ42/Aβ40 ratio should not be altered. However, a dimeric PS at the core of γ-secretase will allow an inactive, cleavage-incompetent PS to alter substrate presentation to and/or the specificity of the WT molecule. In this case, the FAD phenotype will be epistatic to the Asp-385 mutation. FL, full length.
Fig. 3.
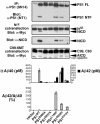
Some PS1 FAD mutations are intrinsically hypomorphic for different proteolytic activities. WT and mutant forms of PS1 were transiently transfected into PS1/PS2-deficient MEFs to characterize their associated activities. To determine PS endoproteolysis, lysates of transfected cells were immunoprecipitated with 9N14, resolved by SDS/PAGE, and immunostained with NT1. To assess NICD production, PS1 was cotransfected with NΔE and 9E10 and α-V1744 antibodies (Cell Signaling Technology, Beverly, MA) were used to detect NICD on Western blots. To determine AICD release, PS1 was cotransfected with C99 – 6MT and AICD was visualized with 9E10 antibody. Aβ production was assessed by ELISA on conditioned media from cells cotransfected with PS1 and APP (n = 4) (see Supporting Methods, which is published as supporting information on the PNAS web site). Average of independent triplicates from one representative experiment is shown. FL, full length.
Similar articles
- Presenilins are required for gamma-secretase cleavage of beta-APP and transmembrane cleavage of Notch-1.
Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner BA. Zhang Z, et al. Nat Cell Biol. 2000 Jul;2(7):463-5. doi: 10.1038/35017108. Nat Cell Biol. 2000. PMID: 10878814 No abstract available. - Notch1 competes with the amyloid precursor protein for gamma-secretase and down-regulates presenilin-1 gene expression.
Lleó A, Berezovska O, Ramdya P, Fukumoto H, Raju S, Shah T, Hyman BT. Lleó A, et al. J Biol Chem. 2003 Nov 28;278(48):47370-5. doi: 10.1074/jbc.M308480200. Epub 2003 Sep 5. J Biol Chem. 2003. PMID: 12960155 - Endoproteolysis of the ER stress transducer ATF6 in the presence of functionally inactive presenilins.
Steiner H, Winkler E, Shearman MS, Prywes R, Haass C. Steiner H, et al. Neurobiol Dis. 2001 Aug;8(4):717-22. doi: 10.1006/nbdi.2001.0405. Neurobiol Dis. 2001. PMID: 11493036 - Implication of APP secretases in notch signaling.
Hartmann D, Tournoy J, Saftig P, Annaert W, De Strooper B. Hartmann D, et al. J Mol Neurosci. 2001 Oct;17(2):171-81. doi: 10.1385/JMN:17:2:171. J Mol Neurosci. 2001. PMID: 11816790 Review. - Presenilin function and gamma-secretase activity.
Brunkan AL, Goate AM. Brunkan AL, et al. J Neurochem. 2005 May;93(4):769-92. doi: 10.1111/j.1471-4159.2005.03099.x. J Neurochem. 2005. PMID: 15857382 Review.
Cited by
- Comparable dimerization found in wildtype and familial Alzheimer's disease amyloid precursor protein mutants.
So PP, Khodr CE, Chen CD, Abraham CR. So PP, et al. Am J Neurodegener Dis. 2013;2(1):15-28. Epub 2013 Mar 8. Am J Neurodegener Dis. 2013. PMID: 23515184 Free PMC article. - Processing of Notch and amyloid precursor protein by gamma-secretase is spatially distinct.
Tarassishin L, Yin YI, Bassit B, Li YM. Tarassishin L, et al. Proc Natl Acad Sci U S A. 2004 Dec 7;101(49):17050-5. doi: 10.1073/pnas.0408007101. Epub 2004 Nov 24. Proc Natl Acad Sci U S A. 2004. PMID: 15563588 Free PMC article. - Inhibitors of γ-secretase stabilize the complex and differentially affect processing of amyloid precursor protein and other substrates.
Barthet G, Shioi J, Shao Z, Ren Y, Georgakopoulos A, Robakis NK. Barthet G, et al. FASEB J. 2011 Sep;25(9):2937-46. doi: 10.1096/fj.11-183806. Epub 2011 May 19. FASEB J. 2011. PMID: 21597003 Free PMC article. - Protein aggregation in the brain: the molecular basis for Alzheimer's and Parkinson's diseases.
Irvine GB, El-Agnaf OM, Shankar GM, Walsh DM. Irvine GB, et al. Mol Med. 2008 Jul-Aug;14(7-8):451-64. doi: 10.2119/2007-00100.Irvine. Mol Med. 2008. PMID: 18368143 Free PMC article. Review. - High resolution imaging study of interactions between the 37 kDa/67 kDa laminin receptor and APP, beta-secretase and gamma-secretase in Alzheimer's disease.
Jovanovic K, Loos B, Da Costa Dias B, Penny C, Weiss SF. Jovanovic K, et al. PLoS One. 2014 Jun 27;9(6):e100373. doi: 10.1371/journal.pone.0100373. eCollection 2014. PLoS One. 2014. PMID: 24972054 Free PMC article.
References
- Selkoe, D. & Kopan, R. (2003) Annu. Rev. Neurosci. 26 565-597. - PubMed
- Haass, C. & Steiner, H. (2002) Trends Cell Biol. 12 556-562. - PubMed
- Annaert, W. & De Strooper, B. (2002) Annu. Rev. Cell Dev. Biol. 18 25-51. - PubMed
- De Strooper, B. & Annaert, W. (2000) J. Cell Sci. 113 1857-1870. - PubMed
- Vassar, R. (2002) Adv. Drug Deliv. Rev. 54 1589-1602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS 41355/NS/NINDS NIH HHS/United States
- GM 55479/GM/NIGMS NIH HHS/United States
- R01 GM055479/GM/NIGMS NIH HHS/United States
- R01 AG017050/AG/NIA NIH HHS/United States
- AG 17050/AG/NIA NIH HHS/United States
- R01 NS041355/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources