Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells - PubMed (original) (raw)
. 2003 Nov;73(5):1092-105.
doi: 10.1086/379523. Epub 2003 Oct 21.
Affiliations
- PMID: 14574643
- PMCID: PMC1180489
- DOI: 10.1086/379523
Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells
Zhi Yang et al. Am J Hum Genet. 2003 Nov.
Abstract
Gene-specific CTG/CAG repeat expansion is associated with at least 14 human diseases, including myotonic dystrophy type 1 (DM1). Most of our understanding of trinucleotide instability is from nonhuman models, which have presented mixed results, supporting replication errors or processes independent of cell division as causes. Nevertheless, the mechanism occurring at the disease loci in patient cells is poorly understood. Using primary fibroblasts derived from a fetus with DM1, we have shown that spontaneous expansion of the diseased (CTG)(216) allele occurred in proliferating cells but not in quiescent cells. Expansions were "synchronous," with mutation frequencies approaching 100%. Furthermore, cells were treated with agents known to alter DNA synthesis but not to directly damage DNA. Inhibiting replication initiation with mimosine had no effect upon instability. Inhibiting both leading- and lagging-strand synthesis with aphidicolin or blocking only lagging strand synthesis with emetine significantly enhanced CTG expansions. It was striking that only the expanded DM1 allele was altered, leaving the normal allele, (CTG)(12), and other repeat loci unaffected. Standard and small-pool polymerase chain reaction revealed that inhibitors enhanced the magnitude of short expansions in most cells threefold, whereas 11%-25% of cells experienced gains of 122-170 repeats, to sizes of (CTG)(338)-(CTG)(386). Similar results were observed for an adult DM1 cell line. Our results support a role for the perturbation of replication fork dynamics in DM1 CTG expansions within patient fibroblasts. This is the first report that repeat-length alterations specific to a disease allele can be modulated by exogenously added compounds.
Figures
Figure 1
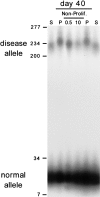
DM1 CTG expansion in cultured DM1 fibroblasts requires cell proliferation. Starting cells (S) were split into control, proliferating cells (P), and growth-arrested cells (Non-Prolif.) regimens. Growth arrest was induced by serum starvation (0.5% serum, indicated by “0.5”) or by confluence-induced contact inhibition (10% serum, indicated by “10”). Proliferating cells were split 1:4 nine times over the 40 d. DNAs were isolated, CTG repeats of the DM1 locus were PCR amplified (see the “Methods” section), and tract lengths were determined on 1.5% agarose gels. The size marker has been converted to repeat numbers. The normal (CTG)12 and expanded diseased (CTG)216 DM1 alleles are indicated. To facilitate comparison, the control starting and proliferating samples were loaded flanking the arrested samples.
Figure 2
Drug treatment and growth of DM1 fibroblasts. A, Strategy of drug treatments. Logarithmically growing cells from the same initial plate were split into control (no drug) and drug treatment regimens. To avoid selection for drug-resistant lines, cells were repeatedly exposed to one drug, with drug-free recovery periods between exposures. One “round” of treatment, including exposure and recovery periods, represented a total of three cell PDs. Cells were maintained between 20% and 80% confluence. Approximately 5–6×105 cells/plate at 40% confluence (100-mm plates) were exposed to 1 μM emetine or 200 μM mimosine for 18 h, during which the rate of replication progressively decreased. Low-dose aphidicolin (0.207 μM) exposure spanned a single PD. After exposure, drugs were removed, and cells were washed with PBS and allowed to recover in fresh, drug-free media. Cells were passed through subsequent treatment/recovery regimens for as many as five rounds of exposure to the same drug. At the end of each recovery period, an aliquot of cells was removed, and genomic DNA was extracted for CTG length analyses. For the “no-drug” controls, an equal amount of cells were mock-treated through the same number of cell doublings, and aliquots were taken for DNA analysis after the same number of PD. B, Proliferative status. Drug exposures occurred between blackened-to-blackened shapes, and drug-free recovery periods occurred between blackened shapes interspersed with unblackened shapes.
Figure 3
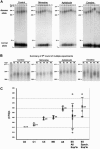
Expansion of chromosomal DM1 CTG repeats in DM1 fibroblasts with or without exposure to replication inhibitors. Control and drug-treated cells were grown through one to five rounds, as outlined in figure 2, and DNAs were isolated for PCR analysis (see the “Methods” section). DM1 CTG repeat lengths were determined on 1.5% agarose gels, relative to a DNA size marker (converted to repeat numbers). A, Results from one set of six independent experiments (experiment #1). PCR products from control cells grown in the absence of drug (starting culture C0, multiple rounds C1–C5). To facilitate length comparisons, flanking each set of PCR products from drug-treated cells are PCR products from control cells (C1 or C5). PCR products are from cells treated with mimosine (rounds M1–M5); aphidicolin (rounds A1–A5); and emetine (rounds E1–E5). B, Summary of the PCR products of fifth round of growth in control, mimosine-, aphidicolin-, and emetine-treated cells from five other independent sets of experiments. The normal DM1 allele remained at (CTG)12 and, for the sake of clarity, only the upper portion of the gel with the unstable disease allele is shown. All panels are portions of the same gel, permitting the comparison between all lanes and the starting C0 sample in the first panel. C, Graphical representation of the sizes of the major diseased CTG products in each fifth round of control and drug-treated samples. For each sample, individual measurements are unblackened symbols, the mean is blackened, and the error bars show plus and minus one sample SD of the data. Two major products were observed in two emetine-treated samples (experiments #4 and #6); hence, there are eight data points for the six repetitions of the experiment. The four emetine-treated samples that displayed synchronous expansions (experiments #1 #2, #3, and #5) are separated out plotted to the right (“Synch.”). The complete sizing of these samples can be found in appendix A (online only).
Figure 4
spPCR analysis of the DM1 CTG tract from control, aphidicolin-treated, and emetine-treated cells. As many as 15–40 individual spPCRs per sample were performed (see the “Methods” section), and CTG lengths of each product in each reaction were determined on 1.8% agarose gels, relative to a DNA size marker (converted to repeat numbers). A, A representative set of spPCR products from the first and fifth rounds of growth of control cells (in the absence of inhibitors) and from the fifth round of growth in the presence of aphidicolin or emetine. The normal DM1 allele remained (CTG)12, and, for the sake of clarity, only the upper portion of the gel with the unstable disease allele is shown. B, Graphical representation of CTG size distributions of individual spPCR products in each sample. To facilitate comparisons, a dashed line indicates the peak CTG size in the fifth round of control cells (C5). The complete sizing and analysis of these and other samples can be found in appendix B (online only).
Figure 5
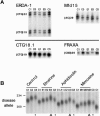
Preferential and actively enhanced expansions of the diseased DM1 CTG repeat. A, Repeat-tract lengths of the ERDA1, CTG18.1, FRAXA, and Mfd15 loci were determined by PCR (see the “Methods” section) in control and drug-treated DM1 cells grown through one to five rounds, as outlined in figure 2. DNAs were isolated for PCR analysis (see the “Methods” section), and tract lengths were determined on denaturing 5% polyacrylamide gels, relative to sequencing ladders. Only the first and fifth rounds of the control and emetine-treated cells are shown. The same analysis was performed on DNA from aphidicolin-treated and mimosine-treated cells; length variation was not observed at any of these repeats (data not shown). B, Expansion of DM1 CTG repeats in a single cell clonal line of the DM1 fibroblasts. Control and drug-treated cells were grown through multiple rounds, and DNAs were isolated as outlined in figure 2_A_. DM1 CTG repeats were PCR amplified, and tract lengths were determined by electrophoresis on 1.5% agarose gels alongside a DNA size marker (converted to repeat numbers). To facilitate length comparisons, flanking each set of PCR products from drug-treated cells are PCR products from control cells (C1 or C3). Although this gel was run at a slant, CTG length differences are observable by comparing flanking C3 with E3 or A3 (compare lanes indicated by asterisks with those indicated by triangles). As indicated, shown are PCR products from control cells grown in the absence of drug (rounds C1–C3), cells treated with emetine (rounds E1–E3), aphidicolin (rounds A1–A3), or mimosine (rounds M1–M3).
Figure 6
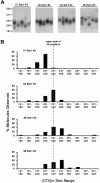
spPCR analysis of the DM1 CTG tract from an adult cell line. As many as 40 individual spPCRs per sample were performed (see the “Methods” section), and CTG lengths of each product in each reaction were determined on 1.8% agarose gels relative to a DNA size marker (converted to repeat numbers). A, A representative set of spPCR products from the first, second, third, fourth, and fifth rounds of growth of control cells grown in the absence of inhibitors (C1–C5). The major and minor populations of cells harboring shorter and longer CTG expansions are indicated as “population 1” and “population 2,” respectively. Individual faint products in population 2 of C1 are indicated by arrows. B, A representative set of spPCR products from the fifth round of growth in the absence (C5) or presence of aphidicolin (A5) or emetine (E5). Panels A and B are portions of the same gel, permitting the comparison between all lanes and the starting C1 samples. C, Graphical representation of CTG size distributions of individual spPCR products in each sample. The complete sizing and analysis of these and other samples can be found in appendix C (online only).
Similar articles
- Oligodeoxynucleotide binding to (CTG) · (CAG) microsatellite repeats inhibits replication fork stalling, hairpin formation, and genome instability.
Liu G, Chen X, Leffak M. Liu G, et al. Mol Cell Biol. 2013 Feb;33(3):571-81. doi: 10.1128/MCB.01265-12. Epub 2012 Nov 19. Mol Cell Biol. 2013. PMID: 23166299 Free PMC article. - Maternal germline-specific effect of DNA ligase I on CTG/CAG instability.
Tomé S, Panigrahi GB, López Castel A, Foiry L, Melton DW, Gourdon G, Pearson CE. Tomé S, et al. Hum Mol Genet. 2011 Jun 1;20(11):2131-43. doi: 10.1093/hmg/ddr099. Epub 2011 Mar 5. Hum Mol Genet. 2011. PMID: 21378394 - Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats.
Nakamori M, Pearson CE, Thornton CA. Nakamori M, et al. Hum Mol Genet. 2011 Feb 1;20(3):580-8. doi: 10.1093/hmg/ddq501. Epub 2010 Nov 18. Hum Mol Genet. 2011. PMID: 21088112 Free PMC article. - Overview of the Complex Relationship between Epigenetics Markers, CTG Repeat Instability and Symptoms in Myotonic Dystrophy Type 1.
de Pontual L, Tomé S. de Pontual L, et al. Int J Mol Sci. 2022 Mar 23;23(7):3477. doi: 10.3390/ijms23073477. Int J Mol Sci. 2022. PMID: 35408837 Free PMC article. Review. - Modifiers of CAG/CTG Repeat Instability: Insights from Mammalian Models.
Wheeler VC, Dion V. Wheeler VC, et al. J Huntingtons Dis. 2021;10(1):123-148. doi: 10.3233/JHD-200426. J Huntingtons Dis. 2021. PMID: 33579861 Free PMC article. Review.
Cited by
- Inhibition of DNA synthesis facilitates expansion of low-complexity repeats: is strand slippage stimulated by transient local depletion of specific dNTPs?
Kuzminov A. Kuzminov A. Bioessays. 2013 Apr;35(4):306-13. doi: 10.1002/bies.201200128. Epub 2013 Jan 15. Bioessays. 2013. PMID: 23319444 Free PMC article. - Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired.
Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Panigrahi GB, et al. Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12593-8. doi: 10.1073/pnas.0909087107. Epub 2010 Jun 22. Proc Natl Acad Sci U S A. 2010. PMID: 20571119 Free PMC article. - Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease.
McMurray CT. McMurray CT. DNA Repair (Amst). 2008 Jul 1;7(7):1121-34. doi: 10.1016/j.dnarep.2008.03.013. Epub 2008 May 9. DNA Repair (Amst). 2008. PMID: 18472310 Free PMC article. Review. - Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile X premutation mouse model.
Entezam A, Lokanga AR, Le W, Hoffman G, Usdin K. Entezam A, et al. Hum Mutat. 2010 May;31(5):611-6. doi: 10.1002/humu.21237. Hum Mutat. 2010. PMID: 20213777 Free PMC article. - MSH3 polymorphisms and protein levels affect CAG repeat instability in Huntington's disease mice.
Tomé S, Manley K, Simard JP, Clark GW, Slean MM, Swami M, Shelbourne PF, Tillier ER, Monckton DG, Messer A, Pearson CE. Tomé S, et al. PLoS Genet. 2013;9(2):e1003280. doi: 10.1371/journal.pgen.1003280. Epub 2013 Feb 28. PLoS Genet. 2013. PMID: 23468640 Free PMC article.
References
Electronic-Database Information
- Fisher’s Exact Test Web site, http://matforsk.no/ola/fisher.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DM1) - PubMed
References
- Anvret M, Ahlberg G, Grandell U, Hedberg B, Johnson K, Edstrom L (1993) Larger expansions of the CTG repeat in muscle compared to lymphocytes from patients with myotonic dystrophy. Hum Mol Genet 2:1397–1400 - PubMed
- Ashizawa T, Monckton DG, Vaishnav S, Patel BJ, Voskova A, Caskey CT (1996) Instability of the expanded (CTG)n repeats in the myotonin protein kinase gene in cultured lymphoblastoid cell lines from patients with myotonic dystrophy. Genomics 36:47–53 - PubMed
- Benitez J, Robledo M, Ramos C, Ayuso C, Astarloa R, Garcia Yebenes J, Brambati B (1995) Somatic stability in chorionic villi samples and other Huntington fetal tissues. Hum Genet 96:229–232 - PubMed
- Breschel TS, McInnis MG, Margolis RL, Sirugo G, Corneliussen B, Simpson SG, McMahon FJ, MacKinnon DF, Xu JF, Pleasant N, Huo Y, Ashworth RG, Grundstrom C, Grundstrom T, Kidd KK, DePaulo JR, Ross CA (1997) A novel, heritable, expanding CTG repeat in an intron of the SEF2–1 gene on chromosome 18q21.1. Hum Mol Genet 6:1855–1863 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources