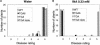Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance - PubMed (original) (raw)
. 2003 Nov;15(11):2647-53.
doi: 10.1105/tpc.014894. Epub 2003 Oct 23.
Affiliations
- PMID: 14576289
- PMCID: PMC280568
- DOI: 10.1105/tpc.014894
Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance
Yuelin Zhang et al. Plant Cell. 2003 Nov.
Abstract
Arabidopsis nonexpresser of pathogenesis-related (PR) genes (NPR1) is the sole positive regulator that has been shown to be essential for the induction of systemic acquired resistance. In npr1 mutant plants, salicylic acid (SA)-mediated PR gene expression and pathogen resistance are abolished completely. NPR1 has been shown to interact with three closely related TGA transcription factors-TGA2, TGA5, and TGA6-in yeast two-hybrid assays. To elucidate the biological functions of these three TGA transcription factors, we analyzed single and combined deletion knockout mutants of TGA2, TGA5, and TGA6 for SA-induced PR gene expression and pathogen resistance. Induction of PR gene expression and pathogen resistance by the SA analog 2,6-dichloroisonicotinic acid (INA) was blocked in tga6-1 tga2-1 tga5-1 but not in tga6-1 or tga2-1 tga5-1 plants. Loss of INA-induced resistance to Peronospora parasitica Noco2 cosegregated with the tga6-1 mutation in progeny of multiple lines that were heterozygous for tga6-1 and homozygous for tga2-1 tga5-1 and could be complemented by genomic clones of wild-type TGA2 or TGA5, indicating that TGA2, TGA5, and TGA6 encode redundant and essential functions in the positive regulation of systemic acquired resistance. In addition, tga6-1 tga2-1 tga5-1 plants had reduced tolerance to high levels of SA and accumulated higher basal levels of PR-1 under noninducing conditions, suggesting that these TGA factors also are important for SA tolerance and the negative regulation of the basal expression of PR-1.
Figures
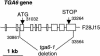
Figure 1.
Structure of the Deletion in tga6-1. The fast neutron–induced deletion in TGA6 spans the region between position 30,861 in the second intron and position 33,564 in the last exon. The numbering is based on the Arabidopsis BAC clone F28J15 that contains TGA6.
Figure 2.
Tolerance of Wild-Type, npr1-1, tga6-1, tga2-1 tga5-1, and tga6-1 tga2-1 tga5-1 Plants to 0.2 mM SA. Seeds were plated on MS medium containing 0.2 mM SA, and the photographs were taken 14 days after germination. This experiment was repeated twice with similar results.
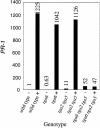
Figure 3.
PR-1 Expression in Wild-Type, tga6-1, tga2-1 tga5-1, and tga6-1 tga2-1 tga5-1 Plants in Response to Treatment with INA. Total RNA was extracted from 20-day-old seedlings grown on MS medium in the presence (+) or absence (−) of 50 μM INA. Relative levels of PR-1 were determined by real-time PCR using SYBR Green I chemistry. Values were normalized to the expression of ACTIN1 and are expressed relative to the level in wild-type plants. This experiment was repeated twice with similar results.
Figure 4.
Growth of P. parasitica Noco2 on Wild-Type, tga6-1, tga2-1 tga5-1, and tga6-1 tga2-1 tga5-1 Plants. Two-week-old seedlings were pretreated with water (A) or 0.33 mM INA (B) and sprayed with P. parasitica Noco2 spores (5 × 103 spores/mL) 3 days later. Infection was scored at 7 days after inoculation by counting the number of conidiophores per infected leaf. A total of 25 plants were scored for each treatment. Disease rating scores are as follows: 0, no conidiophores on the plants; 1, no more than 5 conidiophores per infected leaf; 2, 6 to 20 conidiophores on a few of the infected leaves; 3, 6 to 20 conidiophores on most of the infected leaves; 4, 5 or more conidiophores on all infected leaves; 5, 20 or more conidiophores on all infected leaves. This experiment was repeated once with similar results. WT, wild type.
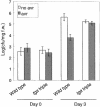
Figure 5.
Growth of P.s.m. ES4326 in Wild-Type and tga6-1 tga2-1 tga5-1 Plants Preinoculated with P.s.t. DC3000 avrRpt2. Two leaves from each plant were infiltrated with P.s.t. DC3000 avrRpt2 (OD600 = 0.02) in 10 mM MgCl2 solution or with the buffer alone 3 days before P.s.m. ES4326 infection (OD600 = 0.001). Leaf discs within the inoculated area were taken at 0 and 3 days after P.s.m. ES4326 infection, and the bacterial titers were measured. Error bars represent 95% confidence limits of log-transformed data. Four samples were taken for each time point. This experiment was repeated once with similar results. avr, P.s.t. DC3000 avrRpt2; cfu, colony-forming units; f.w., fresh weight; tga triple, tga6-1 tga2-1 tga5-1.
Figure 6.
Complementation of tga6-1 tga2-1 tga5-1 by Genomic Clones of TGA2 or TGA5. tga triple, tga6-1 tga2-1 tga5-1; _TGA2_g, a representative tga6-1 tga2-1 tga5-1 line transformed with the genomic clone of TGA2; _TGA5_g, a representative tga6-1 tga2-1 tga5-1 line transformed with the genomic clone of TGA5. (A) Complementation of tga6-1 tga2-1 tga5-1 in PR-1 expression. The relative expression levels of PR-1 in tga6-1 tga2-1 tga5-1, _TGA2_g, and _TGA5_g were determined as described for Figure 3. +, grown on MS medium + 50 mM INA; −, MS medium alone. (B) Complementation of tga6-1 tga2-1 tga5-1 in SA tolerance. T3 seeds of tga6-1 tga2-1 tga5-1 homozygous for the TGA2 or TGA5 transgene were plated on MS medium with 0.2 mM SA along with seeds of tga6-1 tga2-1 tga5-1. The photographs were taken 14 days after gemination. (C) and (D) Complementation of tga6-1 tga2-1 tga5-1 in response to pathogen infection. Using protocols described in Figure 4, water-treated (C) or INA-treated (D) plants were inoculated with P. parasitica Noco2. Infection was scored at 7 days after inoculation by counting the number of conidiophores per infected leaf. A total of 25 plants were scored for each treatment. Each experiment was repeated at least once with similar results.
Similar articles
- Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis.
Kesarwani M, Yoo J, Dong X. Kesarwani M, et al. Plant Physiol. 2007 May;144(1):336-46. doi: 10.1104/pp.106.095299. Epub 2007 Mar 16. Plant Physiol. 2007. PMID: 17369431 Free PMC article. - NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid.
Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. Zhou JM, et al. Mol Plant Microbe Interact. 2000 Feb;13(2):191-202. doi: 10.1094/MPMI.2000.13.2.191. Mol Plant Microbe Interact. 2000. PMID: 10659709 - NPR1: the spider in the web of induced resistance signaling pathways.
Pieterse CM, Van Loon LC. Pieterse CM, et al. Curr Opin Plant Biol. 2004 Aug;7(4):456-64. doi: 10.1016/j.pbi.2004.05.006. Curr Opin Plant Biol. 2004. PMID: 15231270 Review. - Priming in plant-pathogen interactions.
Conrath U, Pieterse CM, Mauch-Mani B. Conrath U, et al. Trends Plant Sci. 2002 May;7(5):210-6. doi: 10.1016/s1360-1385(02)02244-6. Trends Plant Sci. 2002. PMID: 11992826 Review.
Cited by
- A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation.
Chern M, Bai W, Sze-To WH, Canlas PE, Bartley LE, Ronald PC. Chern M, et al. Plant Methods. 2012 Feb 21;8(1):6. doi: 10.1186/1746-4811-8-6. Plant Methods. 2012. PMID: 22353606 Free PMC article. - Genome-wide analysis reveals regulatory mechanisms and expression patterns of TGA genes in peanut under abiotic stress and hormone treatments.
Zhong C, Liu Y, Li Z, Wang X, Jiang C, Zhao X, Kang S, Liu X, Zhao S, Wang J, Zhang H, Huang Y, Yu H, Xue R. Zhong C, et al. Front Plant Sci. 2023 Nov 21;14:1269200. doi: 10.3389/fpls.2023.1269200. eCollection 2023. Front Plant Sci. 2023. PMID: 38078104 Free PMC article. - BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis.
Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. Hepworth SR, et al. Plant Cell. 2005 May;17(5):1434-48. doi: 10.1105/tpc.104.030536. Epub 2005 Apr 1. Plant Cell. 2005. PMID: 15805484 Free PMC article. - CYSTEINE-RICH RECEPTOR-LIKE KINASE5 (CRK5) and CRK22 regulate the response to Verticillium dahliae toxins.
Zhao J, Sun Y, Li X, Li Y. Zhao J, et al. Plant Physiol. 2022 Aug 29;190(1):714-731. doi: 10.1093/plphys/kiac277. Plant Physiol. 2022. PMID: 35674361 Free PMC article. - Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis.
Wang P, Nolan TM, Yin Y, Bassham DC. Wang P, et al. Autophagy. 2020 Jan;16(1):123-139. doi: 10.1080/15548627.2019.1598753. Epub 2019 Apr 6. Autophagy. 2020. PMID: 30909785 Free PMC article.
References
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. - PubMed
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. - PubMed
- Chern, M.S., Fitzgerald, H.A., Yadav, R.C., Canlas, P.E., Dong, X., and Ronald, P.C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27, 101–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous