Histone sumoylation is associated with transcriptional repression - PubMed (original) (raw)
Histone sumoylation is associated with transcriptional repression
Yuzuru Shiio et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Histone proteins are subject to modifications, such as acetylation, methylation, phosphorylation, ubiquitination, glycosylation, and ADP ribosylation, some of which are known to play important roles in the regulation of chromatin structure and function. Here we report that histone H4 is modified by small ubiquitin-related modifier (SUMO) family proteins both in vivo and in vitro. H4 binds to the SUMO-conjugating enzyme (E2), UBC9, and can be sumoylated in an E1 (SUMO-activating enzyme)- and E2-dependent manner. We present evidence suggesting that histone sumoylation mediates gene silencing through recruitment of histone deacetylase and heterochromatin protein 1.
Figures
Fig. 1.
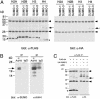
Sumoylation of histone H4 in vivo.(A) Modification of histone H4 by SUMO-1 and SUMO-3. FLAG-tagged histone H2A, H2B, H3, or H4 was cotransfected with HA-tagged SUMO-1 or SUMO-3 (where indicated) into 293T cells. Two days later, anti-FLAG immunoprecipitates (under denaturing conditions) were analyzed by anti-FLAG (Left) or anti-HA (Right) immunoblotting. The asterisks indicate the positions of unmodified histones. The arrow indicates the position of sumoylated H4. The addition of the FLAG tag increases the apparent molecular mass of H4 from 10 to 14 kDa. The apparent molecular mass of the SUMO monomer is ≈15 kDa. Arrowheads indicate Ig chains. The presence of the SUMO moiety in the modified H4 was confirmed by anti-HA immunoblotting (Right). (B) Sumoylation of endogenous histone H4 in human B cells. A chromatin fraction derived from human P493-6 B cells (1 × 108 cells; see Materials and Methods) was immunoprecipitated under denaturing conditions with antiacetylated histone H4 Ab or a control Ab, and the immuno-precipitate was analyzed by anti-SUMO-1 immunoblotting (Left) or antiacetylated histone H4 immunoblotting (Right). The position of sumoylated, acetylated histone H4 is indicated by the arrow. The identity of ≈15- and 18-kDa bands detected by antiacetylated H4 immunoblotting is unknown. The sumoylation of histone H4 appeared to enhance the efficiency of electrotransfer. Most of the nonsumoylated histone H4 remained in the gel after transfer under the conditions used. (C) Enhancement of H4 sumoylation by histone acetyltransferase p300. FLAG-tagged histone H4 was transfected alone or with HA-tagged SUMO-1 or p300 (where indicated) into 293T cells. Two days later, anti-FLAG immunoprecipitates (under denaturing conditions) were analyzed by anti-FLAG (Left) or anti-HA (Right) immunoblotting. The asterisk indicates the positions of unmodified histones. The arrow indicates the position of sumoylated H4. Arrowheads indicate Ig chains. The presence of the SUMO moiety in the modified H4 was confirmed by anti-HA immunoblotting (Right).
Fig. 2.
Sumoylation of histone H4 in vitro. (A) E1- and E2-dependent sumoylation of histone H4. _In vitro_-translated, 35S-Met-labeled histone H4 was incubated with purified E1 (SAE1 0.15 μg/SAE2 0.45 μg), purified E2 (UBC9 0.5 μg), and GST-SUMO-1 (0.5 μg) (where indicated) at 30°C for 3 h. The positions of unmodified H4 and H4 modified with GST-SUMO-1 are indicated by arrows. (B) Sumoylation of the N-terminal tail of histone H4. _In vitro-_translated, 35S-Met-labeled full-length histone H4 or the N-terminal tail of H4 (H4N, amino acid 1–26) was subjected to the sumoylation reaction with GST-SUMO-1, or GST-SUMO-3, and E1 and E2 as in A. The positions of modified H4 or H4N are indicated by asterisks. An aliquot of the sumoylation reaction mixture was bound to glutathione agarose beads to assess the association of GST-SUMO proteins with H4. (C) Binding of histone H4 to UBC9. _In vitro-_translated, 35S-Met-labeled histone H4 was incubated with GST, GST-UBC9, GST-SUMO-1, or GST-SUMO-3 attached to glutathione agarose beads. The bound proteins were analyzed by SDS/PAGE (Left). As a positive control, the interaction of RanGAP1 with GST-UBC9 was also examined (Right). The input lanes represent 10% of the input to the binding reactions.
Fig. 3.
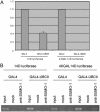
Chromatin sumoylation is associated with transcriptional repression by GAL4-UBC9. (A) Transcriptional repression by GAL4-UBC9. HeLa cells were cotransfected with 2 μg of 4XGAL14D luciferase reporter and indicated amount of GAL4, GAL4-UBC9 (full-length, amino acid 1–158), GAL4-UBC9ΔN (35–158), or GAL4-UBC9ΔC(1–92). After transfection (48 h), the luciferase activities were determined. The data represent the average of two independent experiments. Comparable expression of GAL4, GAL4-UBC9, GAL4-UBC9ΔN, and GAL4-UBC9ΔC was confirmed by anti-GAL4 immunoblotting (data not shown). (B) Chromatin sumoylation by GAL4-UBC9. HeLa cells were cotransfected with 4XGAL14D luciferase reporter and GAL4 or GAL4-UBC9. Two days later, sumoylation of chromatin near the GAL4 site was examined by anti-SUMO-1 chromatin immunoprecipitation. Mock lanes represent the chromatin immunoprecipitation without the Ab. Chromatin sumoylation by GAL4-UBC9 was detected by chromatin immunoprecipitation under both nondenaturing (Left) and denaturing (Right) conditions. (C) SUMO-conjugating activities of UBC9 deletion mutants. The 293T cells were cotransfected with HA-SUMO-1 or HA-SUMO-3 together with a vector, UBC9, UBC9DN, or UBC9DC. Whole-cell lysate (30 μg) was analyzed by anti-HA immunoblotting for sumoylation of cellular proteins.
Fig. 4.
Transcriptional repression and chromatin sumoylation by GAL4-UBC9 depend on the GAL4-binding sites. (A) Transcriptional repression by GAL4-UBC9 depends on the GAL4-binding sites. HeLa cells were cotransfected with 2 μg of 14D luciferase reporter (without GAL4-binding sites) or 4XGAL14D luciferase reporter (with GAL4-binding sites) and 5 μg of GAL4 or GAL4-UBC9. Luciferase activities were determined 48 h after transfection. The data shown represent the average of two independent experiments. (B) Chromatin sumoylation by GAL4-UBC9 depends on the presence of GAL4-binding sites. HeLa cells were cotransfected with 14D luciferase reporter (without GAL4-binding sites) or 4XGAL14D luciferase reporter (with GAL4-binding sites) and GAL4 or GAL4-UBC9. Chromatin sumoylation by GAL4-UBC9 was detected by chromatin immunoprecipitation under nondenaturing condition as in Fig. 3_B_.
Fig. 5.
Histone sumoylation is associated with recruitment of HDAC and HP1. (A) Incorporation of SUMO-H4 fusion proteins into chromatin. The 293 cells were transfected with FLAG-vector (FV), FLAG-H4, FLAG-SUMO-1-H4, FLAG-SUMO-3-H4, FLAG-SUMO-1, or FLAG-SUMO-3, and the cells were fractionated into soluble (S) and chromatin (C) fractions as described (26). The location of each FLAG-tagged protein was determined by anti-FLAG immunoblotting. (B) Association of SUMO-H4 fusion proteins with core histones. The 293 cells were transfected with FLAG vector, FLAG-H4, FLAG-SUMO-1-H4, FLAG-SUMO-3-H4, FLAG-SUMO-1, or FLAG-SUMO-3, and 2 days later cell lysates were immunoprecipitated with anti-FLAG Ab under nondenaturing conditions. The immunoprecipitates were analyzed by SDS/PAGE and Coomassie staining for the presence of core histones (H4, H2A/B, and H3). (C) Interaction of SUMO-H4 fusion proteins with HDAC1 and HP1-γ. The 293 cells were transfected with FLAG vector, FLAG-H4, FLAG-SUMO-1-H4, FLAG-SUMO-3-H4, FLAG-SUMO-1, or FLAG-SUMO-3, and 2 days later cell lysates were immunoprecipitated with anti-FLAG Ab (under native conditions) and analyzed by anti-HDAC1 (Top) or anti-HP1-γ (Middle) immunoblotting. The expression level of each FLAG-tagged protein (shown by *) was examined by anti-FLAG immunoblotting (Bottom). The position of Ig light chain is shown by the arrowhead (Bottom). (D) Histone deacetylation and recruitment of HP1-γ induced by GAL4-UBC9. HeLa cells were cotransfected with 4XGAL14D luciferase reporter and GAL4 or GAL4-UBC9. Two days later, histone acetylation status and recruitment of HP1-γ near the GAL4 sites were examined by antiacetylated histone H3 (Left) and anti-HP1-γ (Right) chromatin immunoprecipitation under nondenaturing conditions.
Comment in
- Histone modifications: Now summoning sumoylation.
Nathan D, Sterner DE, Berger SL. Nathan D, et al. Proc Natl Acad Sci U S A. 2003 Nov 11;100(23):13118-20. doi: 10.1073/pnas.2436173100. Epub 2003 Nov 3. Proc Natl Acad Sci U S A. 2003. PMID: 14597707 Free PMC article. No abstract available.
Similar articles
- Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications.
Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Zhao X, et al. Mol Cell Biol. 2005 Oct;25(19):8456-64. doi: 10.1128/MCB.25.19.8456-8464.2005. Mol Cell Biol. 2005. PMID: 16166628 Free PMC article. - Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription.
Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. Ling Y, et al. Nucleic Acids Res. 2004 Jan 29;32(2):598-610. doi: 10.1093/nar/gkh195. Print 2004. Nucleic Acids Res. 2004. PMID: 14752048 Free PMC article. - Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex.
Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S. Rampalli S, et al. Mol Cell Biol. 2005 Oct;25(19):8415-29. doi: 10.1128/MCB.25.19.8415-8429.2005. Mol Cell Biol. 2005. PMID: 16166625 Free PMC article. - SUMO association with repressor complexes, emerging routes for transcriptional control.
Garcia-Dominguez M, Reyes JC. Garcia-Dominguez M, et al. Biochim Biophys Acta. 2009 Jun-Aug;1789(6-8):451-9. doi: 10.1016/j.bbagrm.2009.07.001. Epub 2009 Jul 17. Biochim Biophys Acta. 2009. PMID: 19616654 Review. - Transcriptional regulation by C-terminal binding proteins.
Chinnadurai G. Chinnadurai G. Int J Biochem Cell Biol. 2007;39(9):1593-607. doi: 10.1016/j.biocel.2007.01.025. Epub 2007 Feb 4. Int J Biochem Cell Biol. 2007. PMID: 17336131 Review.
Cited by
- Understanding the Mechanisms by Which Epigenetic Modifiers Avert Therapy Resistance in Cancer.
Quagliano A, Gopalakrishnapillai A, Barwe SP. Quagliano A, et al. Front Oncol. 2020 Jun 24;10:992. doi: 10.3389/fonc.2020.00992. eCollection 2020. Front Oncol. 2020. PMID: 32670880 Free PMC article. Review. - Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes.
Liu HW, Zhang J, Heine GF, Arora M, Gulcin Ozer H, Onti-Srinivasan R, Huang K, Parvin JD. Liu HW, et al. Nucleic Acids Res. 2012 Nov 1;40(20):10172-86. doi: 10.1093/nar/gks819. Epub 2012 Aug 31. Nucleic Acids Res. 2012. PMID: 22941651 Free PMC article. - K4, K9 and K18 in human histone H3 are targets for biotinylation by biotinidase.
Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. Kobza K, et al. FEBS J. 2005 Aug;272(16):4249-59. doi: 10.1111/j.1742-4658.2005.04839.x. FEBS J. 2005. PMID: 16098205 Free PMC article. - Structural and sequence motifs of protein (histone) methylation enzymes.
Cheng X, Collins RE, Zhang X. Cheng X, et al. Annu Rev Biophys Biomol Struct. 2005;34:267-94. doi: 10.1146/annurev.biophys.34.040204.144452. Annu Rev Biophys Biomol Struct. 2005. PMID: 15869391 Free PMC article. Review. - SUMO modification regulates the transcriptional activity of XBP1.
Chen H, Qi L. Chen H, et al. Biochem J. 2010 Jul 1;429(1):95-102. doi: 10.1042/BJ20100193. Biochem J. 2010. PMID: 20408817 Free PMC article.
References
- Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41-45. - PubMed
- Berger, S. L. (2001) Oncogene 20, 3007-3013. - PubMed
- Kuo, M. H. & Allis, C. D. (1998) BioEssays 20, 615-626. - PubMed
- Melchior, F. (2000) Annu. Rev. Cell Dev. Biol. 16, 591-626. - PubMed
- Hay, R. T. (2001) Trends Biochem. Sci. 26, 332-333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous