Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data - PubMed (original) (raw)
Review
Diagnostic and prognostic prediction using gene expression profiles in high-dimensional microarray data
R Simon. Br J Cancer. 2003.
Abstract
DNA microarrays are a potentially powerful technology for improving diagnostic classification, treatment selection and therapeutics development. There are, however, many potential pitfalls in the use of microarrays that result in false leads and erroneous conclusions. This paper provides a review of the key features to be observed in developing diagnostic and prognostic classification systems based on gene expression profiling and some of the pitfalls to be aware of in reading reports of microarray-based studies.
Figures
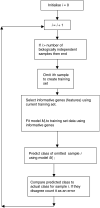
Figure 1
Schematic diagram of leave-one-out cross-validation (LOOCV).
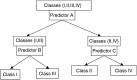
Figure 2
Tree structured classifier for predicting the unknown class of a tissue specimen when four classes are possible. Binary classifier A based on gene expression profile is first used to predict whether the specimen is in subset {I,III} or in subset {II,IV} of classes. For those specimens predicted to be in subset {I,III}, binary classifier B is used to predict whether the specimen is in class I or III. For those specimens predicted to be in subset {II,IV} based on classifier A, binary classifier C is used to predict whether the specimen is in class II or IV. The three binary classifiers will generally utilise different gene sets for prediction.
Similar articles
- Using DNA microarrays for diagnostic and prognostic prediction.
Simon R. Simon R. Expert Rev Mol Diagn. 2003 Sep;3(5):587-95. doi: 10.1586/14737159.3.5.587. Expert Rev Mol Diagn. 2003. PMID: 14510179 Review. - Applications of array technology: melanoma research and diagnosis.
Nambiar S, Mirmohammadsadegh A, Bär A, Bardenheuer W, Roeder G, Hengge UR. Nambiar S, et al. Expert Rev Mol Diagn. 2004 Jul;4(4):549-57. doi: 10.1586/14737159.4.4.549. Expert Rev Mol Diagn. 2004. PMID: 15225102 Review. - Microarrays for cancer diagnosis and classification.
Perez-Diez A, Morgun A, Shulzhenko N. Perez-Diez A, et al. Adv Exp Med Biol. 2007;593:74-85. doi: 10.1007/978-0-387-39978-2_8. Adv Exp Med Biol. 2007. PMID: 17265718 Review. - Converting a breast cancer microarray signature into a high-throughput diagnostic test.
Glas AM, Floore A, Delahaye LJ, Witteveen AT, Pover RC, Bakx N, Lahti-Domenici JS, Bruinsma TJ, Warmoes MO, Bernards R, Wessels LF, Van't Veer LJ. Glas AM, et al. BMC Genomics. 2006 Oct 30;7:278. doi: 10.1186/1471-2164-7-278. BMC Genomics. 2006. PMID: 17074082 Free PMC article. - [Microarray data analysis].
Borup RH, Csillag C, Nielsen OH, Nielsen FC. Borup RH, et al. Ugeskr Laeger. 2006 May 29;168(22):2159-62. Ugeskr Laeger. 2006. PMID: 16768955 Danish.
Cited by
- RiGoR: reporting guidelines to address common sources of bias in risk model development.
Kerr KF, Meisner A, Thiessen-Philbrook H, Coca SG, Parikh CR. Kerr KF, et al. Biomark Res. 2015 Jan 24;3(1):2. doi: 10.1186/s40364-014-0027-7. eCollection 2015. Biomark Res. 2015. PMID: 25642328 Free PMC article. - High-resolution mapping of genomic imbalance and identification of gene expression profiles associated with differential chemotherapy response in serous epithelial ovarian cancer.
Bernardini M, Lee CH, Beheshti B, Prasad M, Albert M, Marrano P, Begley H, Shaw P, Covens A, Murphy J, Rosen B, Minkin S, Squire JA, Macgregor PF. Bernardini M, et al. Neoplasia. 2005 Jun;7(6):603-13. doi: 10.1593/neo.04760. Neoplasia. 2005. PMID: 16036111 Free PMC article. - Competence Classification of Cumulus and Granulosa Cell Transcriptome in Embryos Matched by Morphology and Female Age.
Borup R, Thuesen LL, Andersen CY, Nyboe-Andersen A, Ziebe S, Winther O, Grøndahl ML. Borup R, et al. PLoS One. 2016 Apr 29;11(4):e0153562. doi: 10.1371/journal.pone.0153562. eCollection 2016. PLoS One. 2016. PMID: 27128483 Free PMC article. - The use of evidence from high-throughput screening and transcriptomic data in human health risk assessments.
Mezencev R, Subramaniam R. Mezencev R, et al. Toxicol Appl Pharmacol. 2019 Oct 1;380:114706. doi: 10.1016/j.taap.2019.114706. Epub 2019 Aug 7. Toxicol Appl Pharmacol. 2019. PMID: 31400414 Free PMC article. - Enhancing interdisciplinary mathematics and biology education: a microarray data analysis course bridging these disciplines.
Tra YV, Evans IM. Tra YV, et al. CBE Life Sci Educ. 2010 Fall;9(3):217-26. doi: 10.1187/cbe.09-09-0067. CBE Life Sci Educ. 2010. PMID: 20810954 Free PMC article.
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrich JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511 - PubMed
- Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z (2000) Tissue classification with gene expression profiles. J Comput Biol 7: 559–584 - PubMed
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, VSondak Hayward N, Trent J (2000) Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 406: 536–540 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources