Purification and characterization of the PcrA helicase of Bacillus anthracis - PubMed (original) (raw)
Purification and characterization of the PcrA helicase of Bacillus anthracis
Asma Naqvi et al. J Bacteriol. 2003 Nov.
Abstract
PcrA is an essential helicase in gram-positive bacteria, and a gene encoding this helicase has been identified in all such organisms whose genomes have been sequenced so far. The precise role of PcrA that makes it essential for cell growth is not known; however, PcrA does not appear to be necessary for chromosome replication. The pcrA gene was identified in the genome of Bacillus anthracis on the basis of its sequence homology to the corresponding genes of Bacillus subtilis and Staphylococcus aureus, with which it shares 76 and 72% similarity, respectively. The pcrA gene of B. anthracis was isolated by PCR amplification and cloning into Escherichia coli. The PcrA protein was overexpressed with a His6 fusion at its amino-terminal end. The purified His-PcrA protein showed ATPase activity that was stimulated in the presence of single-stranded (ss) DNA (ssDNA). Interestingly, PcrA showed robust 3'-->5' as well as 5'-->3' helicase activities, with substrates containing a duplex region and a 3' or 5' ss poly(dT) tail. PcrA also efficiently unwound oligonucleotides containing a duplex region and a 5' or 3' ss tail with the potential to form a secondary structure. DNA binding experiments showed that PcrA bound much more efficiently to oligonucleotides containing a duplex region and a 5' or 3' ss tail with a potential to form a secondary structure than to those with ssDNAs or duplex DNAs with ss poly(dT) tails. Our results suggest that specialized DNA structures and/or sequences represent natural substrates of PcrA in biochemical processes that are essential for the growth and survival of gram-positive organisms, including B. anthracis.
Figures
FIG. 1.
Alignment of PcrA of B. anthracis (Ba), B. stearothermophilus (Bst), B. subtilis (Bs), and S. aureus (Sa) in the seven conserved helicase motifs. Amino acids that differ from those of B. anthracis PcrA are shaded and boxed. Numbers correspond to the amino acid positions of the B. anthracis PcrA.
FIG. 2.
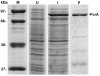
SDS-PAGE analysis of the purified B. anthracis PcrA protein. U, lysates from uninduced cells; I, lysates from IPTG-induced cells overexpressing the His-PcrA protein; P, PcrA protein purified by nickel affinity column chromatography; M, protein molecular-weight standards (in kilodaltons). Smaller fragments likely correspond to breakdown products of PcrA.
FIG. 3.
ATPase activity of PcrA. (A) Products of [α-32P]dATP hydrolysis in the presence of increasing amounts of PcrA. (B) Stimulation of the dATPase activity of PcrA by a 53mer ss oligonucleotide (ssDNA). The products of [α-32P]dATP hydrolysis were analyzed by thin-layer chromatography.
FIG. 4.
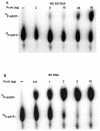
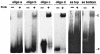
Helicase activity of the PcrA protein. 32P-labeled substrates were incubated with the indicated amounts of PcrA, and the products were resolved by native polyacrylamide gel electrophoresis. The probes used are listed in Table 1 and correspond to duplex oligonucleotides (oligo) containing either a 5′ or 3′ ss region. Only one strand of the probe was labeled. “Complex” corresponds to a PcrA-DNA complex.
FIG. 5.
Binding of the PcrA helicase to various DNA substrates. The PcrA helicase (200 ng) was incubated with probes labeled at the 5′ end, and the DNA-protein complexes were resolved by electrophoresis on native 6% polyacrylamide gels. The probes used are indicated. ss top, top strand of oligonucleotide d; ss bottom, bottom strand of oligonucleotide c. P, free probe; C, PcrA-DNA complex.
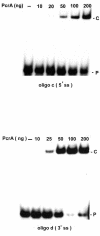
FIG. 6.
Dose-dependent binding of PcrA to duplex DNA substrates containing 5′ or 3′ ss regions (oligonucleotide [oligo] c or oligonucleotide d, respectively). 32P-labeled oligonucleotides were incubated with the indicated amounts of PcrA, and the products were resolved by polyacrylamide gel electrophoresis. P, free probe; C, PcrA-DNA complex.
Similar articles
- Structure-specific DNA binding and bipolar helicase activities of PcrA.
Anand SP, Khan SA. Anand SP, et al. Nucleic Acids Res. 2004 Jun 15;32(10):3190-7. doi: 10.1093/nar/gkh641. Print 2004. Nucleic Acids Res. 2004. PMID: 15199167 Free PMC article. - Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus.
Anand SP, Mitra P, Naqvi A, Khan SA. Anand SP, et al. J Bacteriol. 2004 Apr;186(7):2195-9. doi: 10.1128/JB.186.7.2195-2199.2004. J Bacteriol. 2004. PMID: 15028705 Free PMC article. - PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication.
Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C. Petit MA, et al. Mol Microbiol. 1998 Jul;29(1):261-73. doi: 10.1046/j.1365-2958.1998.00927.x. Mol Microbiol. 1998. PMID: 9701819 - Escherichia coli ribosomal protein L3 stimulates the helicase activity of the Bacillus stearothermophilus PcrA helicase.
Soultanas P, Dillingham MS, Wigley DB. Soultanas P, et al. Nucleic Acids Res. 1998 May 15;26(10):2374-9. doi: 10.1093/nar/26.10.2374. Nucleic Acids Res. 1998. PMID: 9580688 Free PMC article. - Genetic and biochemical characterization of the Streptococcus pneumoniae PcrA helicase and its role in plasmid rolling circle replication.
Ruiz-Masó JA, Anand SP, Espinosa M, Khan SA, del Solar G. Ruiz-Masó JA, et al. J Bacteriol. 2006 Nov;188(21):7416-25. doi: 10.1128/JB.01010-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936036 Free PMC article.
Cited by
- A pathogenicity locus of Streptococcus gallolyticus subspecies gallolyticus.
Taylor JC, Kumar R, Xu J, Xu Y. Taylor JC, et al. Sci Rep. 2023 Apr 18;13(1):6291. doi: 10.1038/s41598-023-33178-z. Sci Rep. 2023. PMID: 37072463 Free PMC article. - Molecular insights into replication initiation in a multipartite genome harboring bacterium Deinococcus radiodurans.
Maurya GK, Chaudhary R, Pandey N, Misra HS. Maurya GK, et al. J Biol Chem. 2021 Jan-Jun;296:100451. doi: 10.1016/j.jbc.2021.100451. Epub 2021 Feb 21. J Biol Chem. 2021. PMID: 33626388 Free PMC article. - Helicases as molecular motors: An insight.
Tuteja N, Tuteja R. Tuteja N, et al. Physica A. 2006 Dec 1;372(1):70-83. doi: 10.1016/j.physa.2006.05.014. Epub 2006 Jun 5. Physica A. 2006. PMID: 32288077 Free PMC article. - Structural and mechanistic insight into DNA unwinding by Deinococcus radiodurans UvrD.
Stelter M, Acajjaoui S, McSweeney S, Timmins J. Stelter M, et al. PLoS One. 2013 Oct 15;8(10):e77364. doi: 10.1371/journal.pone.0077364. eCollection 2013. PLoS One. 2013. PMID: 24143224 Free PMC article. - Functional analysis of the superfamily 1 DNA helicases encoded by Mycoplasma pneumoniae and Mycoplasma genitalium.
Estevão S, van der Heul HU, Sluijter M, Hoogenboezem T, Hartwig NG, van Rossum AM, Vink C. Estevão S, et al. PLoS One. 2013 Jul 23;8(7):e70870. doi: 10.1371/journal.pone.0070870. Print 2013. PLoS One. 2013. PMID: 23894687 Free PMC article.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl. 1988. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- Bird, L. E., S. Subramanya, and D. B. Wigley. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14-18. - PubMed
- Bruand, C., and S. D. Ehrlich. 2000. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol. 35:204-210. - PubMed
- Bruck, I., and M. O'Donnell. 2000. The DNA replication machine of a gram-positive organism. J. Biol. Chem. 275:28971-28983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases