RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria - PubMed (original) (raw)
RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria
Jörg Vogel et al. Nucleic Acids Res. 2003.
Abstract
Recent bioinformatics-aided searches have identified many new small RNAs (sRNAs) in the intergenic regions of the bacterium Escherichia coli. Here, a shot-gun cloning approach (RNomics) was used to generate cDNA libraries of small sized RNAs. Besides many of the known sRNAs, we found new species that were not predicted previously. The present work brings the number of sRNAs in E.coli to 62. Experimental transcription start site mapping showed that some sRNAs were encoded from independent genes, while others were processed from mRNA leaders or trailers, indicative of a parallel transcriptional output generating sRNAs co-expressed with mRNAs. Two of these RNAs (SroA and SroG) consist of known (THI and RFN) riboswitch elements. We also show that two recently identified sRNAs (RyeB and SraC/RyeA) interact, resulting in RNase III-dependent cleavage. To the best of our knowledge, this represents the first case of two non-coding RNAs interacting by a putative antisense mechanism. In addition, intracellular metabolic stabilities of sRNAs were determined, including ones from previous screens. The wide range of half-lives (<2 to >32 min) indicates that sRNAs cannot generally be assumed to be metabolically stable. The experimental characterization of sRNAs analyzed here suggests that the definition of an sRNA is more complex than previously assumed.
Figures
Figure 1
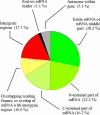
Fractions of the final contigs listed in the Supplementary Material Table S1 with regard to their origin in coding or intergenic regions.
Figure 2
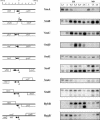
Genomic location and expression patterns of sRNAs. Left: location of sRNA region and neighboring genes (above line, + strand; below line, – strand). Thin vertical bars represent the mapped processed end of sRNA; the thick vertical bars and ‘+1’ represent the mapped 5′ triphosphate end (transcription initiation site; TSS). Right: autoradiograms of northern experiments. Time points represent hours of growth after 100-fold dilution of overnight cultures in LB or M9 medium (Materials and Methods). End-labeled MspI-digested pUC19 DNA was run as a size marker (not shown). SroA: the gene name thiB instead of tbpA was used in accordance with Rodionov et al. (26) and Webb et al. (27).
Figure 3
RACE mapping of sRNA 5′ ends. Specific or increased PCR signals, upon treatment with tobacco acid pyrophosphatase (lane +) compared with mock treatment (lane –), are shown by filled triangles and indicate 5′ triphosphate RNA ends. Open triangles: processed RNA 5′ ends. Other control samples included untreated, 5′ phosphorylated and total DNA (data not shown).
Figure 4
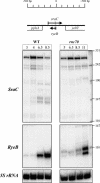
RyeB- and RNaseIII-dependent processing of SraC. Total RNA from isogenic strains (rnc+ and rnc70) was analyzed by northern blot as in Figure 1. Lower panel: autoradiograms of membranes successively probed for SraC, RyeB and, as loading control, 5S rRNA. Upper panel: location of sraC and ryeB in pphA-yebY IGR.
Figure 5
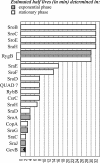
Stability of new sRNAs. Half-lives (in min) were determined in rifampicin run-out experiments in exponentially growing cells (filled bars) or stationary phase (open bars). QUAD denotes probing with an oligodeoxyribonucleotide complementary to sRNA genes in QUAD repeats 1a, 1b and 1c (59), identified as ryeC, ryeD and rygD in Wassarman et al. (6).
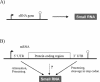
Figure 6
Definitions of sRNAs. (A) Origin of a distinct sRNA species in an own sRNA gene or (B) through parallel transcriptional output, in an mRNA gene.
Similar articles
- How to find small non-coding RNAs in bacteria.
Vogel J, Sharma CM. Vogel J, et al. Biol Chem. 2005 Dec;386(12):1219-38. doi: 10.1515/BC.2005.140. Biol Chem. 2005. PMID: 16336117 Review. - Regulation of RyeA/SraC expression in Escherichia coli.
Gupta AK, Siddiqui N, Yadav D, Arora L, Dutta T. Gupta AK, et al. Biochem Biophys Res Commun. 2019 Aug 27;516(3):661-665. doi: 10.1016/j.bbrc.2019.06.110. Epub 2019 Jun 24. Biochem Biophys Res Commun. 2019. PMID: 31248592 - sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes.
Livny J, Fogel MA, Davis BM, Waldor MK. Livny J, et al. Nucleic Acids Res. 2005 Jul 26;33(13):4096-105. doi: 10.1093/nar/gki715. Print 2005. Nucleic Acids Res. 2005. PMID: 16049021 Free PMC article. - Emergence of New sRNAs in Enteric Bacteria is Associated with Low Expression and Rapid Evolution.
Kacharia FR, Millar JA, Raghavan R. Kacharia FR, et al. J Mol Evol. 2017 Apr;84(4):204-213. doi: 10.1007/s00239-017-9793-9. Epub 2017 Apr 12. J Mol Evol. 2017. PMID: 28405712 - Identification of bacterial small non-coding RNAs: experimental approaches.
Altuvia S. Altuvia S. Curr Opin Microbiol. 2007 Jun;10(3):257-61. doi: 10.1016/j.mib.2007.05.003. Epub 2007 Jun 5. Curr Opin Microbiol. 2007. PMID: 17553733 Review.
Cited by
- Role of Small Non-Coding RNA in Gram-Negative Bacteria: New Insights and Comprehensive Review of Mechanisms, Functions, and Potential Applications.
Khaledi M, Khatami M, Hemmati J, Bakhti S, Hoseini SA, Ghahramanpour H. Khaledi M, et al. Mol Biotechnol. 2024 Aug 17. doi: 10.1007/s12033-024-01248-w. Online ahead of print. Mol Biotechnol. 2024. PMID: 39153013 Review. - Unraveling the interplay between a small RNA and RNase E in bacteria.
Vigoda MB, Argaman L, Kournos M, Margalit H. Vigoda MB, et al. Nucleic Acids Res. 2024 Aug 27;52(15):8947-8966. doi: 10.1093/nar/gkae621. Nucleic Acids Res. 2024. PMID: 39036964 Free PMC article. - A novel ionizing radiation-induced small RNA, DrsS, promotes the detoxification of reactive oxygen species in Deinococcus radiodurans.
Rai SN, Dutta T. Rai SN, et al. Appl Environ Microbiol. 2024 May 21;90(5):e0153823. doi: 10.1128/aem.01538-23. Epub 2024 Apr 8. Appl Environ Microbiol. 2024. PMID: 38587394 Free PMC article. - Directed Screening for sRNA Targets in E. coli Using a Plasmid Library.
Luo X, Majdalani N. Luo X, et al. Methods Mol Biol. 2024;2741:291-306. doi: 10.1007/978-1-0716-3565-0_16. Methods Mol Biol. 2024. PMID: 38217660 - Comparative Analysis of Transcriptome and Proteome Revealed the Common Metabolic Pathways Induced by Prevalent ESBL Plasmids in Escherichia coli.
Huang C, Pham HQ, Zhu L, Wang R, Law OK, Lin SL, Nie QC, Zhang L, Wang X, Lau TC. Huang C, et al. Int J Mol Sci. 2023 Sep 12;24(18):14009. doi: 10.3390/ijms241814009. Int J Mol Sci. 2023. PMID: 37762311 Free PMC article.
References
- Wagner E.G.H. and Vogel,J. (2003) Noncoding RNAs encoded by bacterial chromosomes. In Barciszewski,J. and Erdmann,V. (eds), Noncoding RNAs. Landes Bioscience, pp. 243–259.
- Wassarman K.M., Zhang,A. and Storz,G. (1999) Small RNAs in Escherichia coli. Trends Microbiol., 7, 37–45. - PubMed
- Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. - PubMed
- Argaman L., Hershberg,R., Vogel,J., Bejerano,G., Wagner,E.G.H., Margalit,H. and Altuvia,S. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol., 11, 941–950. - PubMed
- Rivas E., Klein,R.J., Jones,T.A. and Eddy,S.R. (2001) Computational identification of noncoding RNAs in E.coli by comparative genomics. Curr. Biol., 11, 1369–1373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases