The outs and ins of bacterial type IV secretion substrates - PubMed (original) (raw)
Review
The outs and ins of bacterial type IV secretion substrates
Zhiyong Ding et al. Trends Microbiol. 2003 Nov.
Abstract
Bacteria use type IV secretion systems (T4SS) to translocate macromolecular substrates destined for bacterial, plant or human target cells. The T4SS are medically important, contributing to virulence-gene spread, genome plasticity and the alteration of host cellular processes during infection. The T4SS are ancestrally related to bacterial conjugation machines, but present-day functions include (i) conjugal transfer of DNA by cell-to-cell contact, (ii) translocation of effector molecules to eukaryotic target cells, and (iii) DNA uptake from or release to the extracellular milieu. Rapid progress has been made toward identification of type IV secretion substrates and the requirements for substrate recognition.
Figures
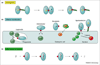
Figure 1
The type IV secretion systems (T4SS) translocate DNA and protein substrates by cell-contact-dependent and –independent mechanisms. Three functional subfamilies include the (a) conjugation systems composed of a transfer channel (red trapezoid) and, for Gram-negative bacteria, an extracellular pilus, (b) ‘effector translocators’ dedicated to the transfer of effector molecules during infection, and (c) DNA uptake or release systems that translocate DNA independently of target-cell contact. Thick and thin squiggly lines are double- and single-stranded DNA, respectively.
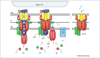
Figure 2
Conservation of type IV secretion genes. The Agrobacterium tumefaciens VirB–D4 reference system is a type IVA secretion system composed of 11 VirB proteins (Mpf) and the VirD4 T4CP (type IV coupling protein), whereas the Dot–Icm, R64 and ColIb-P9 systems are representatives of the type IVB subclass. Dot–Icm proteins discussed in the text or related to VirB proteins are identified. Genes encoding protein homologs are shown. These are not necessarily in the order found in the respective genomes. Homologs of the VirB4 ATPase and VirB7 through to VirB10 are postulated to comprise an ancestral ‘core’ structure to which function-specifying subunits or protein subassemblies were added to evolve the present-day family.
Figure 3
Processing and recruitment of the VirE2 effector to the Agrobacterium tumefaciens VirB–D4 system. Top: Newly synthesized VirE2 forms a stabilizing complex with the secretion chaperone VirE1. Chaperone interactions with N-terminal and central domains prevent VirE2 self-aggregation and formation of premature complexes with other exported effectors, for example, the T-strand. A secretion signal located near the C-terminus of VirE2 mediates complex formation with the VirD4 T4CP (type IV coupling protein) without contributions from VirE1 or the VirB proteins. Lower: Genetic requirements for complex formation between VirE1 and VirE2, and between VirE2 and VirD4, as defined with novel cytology two-hybrid (C2H) and bimolecular fluorescence complementation (BiFC) assays. Fluorescence patterns of cells shown result from production of full- or half-length GFP fusion proteins listed below each image, and the patterns supply evidence for the interactions depicted above each image. Green circle, GFP; purple circle, DivIVA.
Figure 4
Possible architectures and substrate translocation routes for the type IV systems. A Mpf (mating pair formation) core structure might function as a competence machine with only minor evolutionary adaptation. Correspondingly, the addition of inner membrane translocases and, for some systems, a pilus biogenesis pathway, yields the DNA and protein translocation systems at the left. Three working models describe the possible machine architectures and translocation routes: (1) a one-step model using a transenvelope channel, (2) a two-step model using the T4CP or alternative translocase for subtrate transfer across the inner membrane and the Mpf complex for outer membrane transit, and (3) a different two-step model, the ‘shoot and pump’ model, whereby the T4CP recruits substrates and translocates DNA across the inner membrane, and delivers protein substrates to the Mpf protein export apparatus. Blue line, T-strand substrate; red circle, relaxase bound to the T-strand; green circle, protein substrate.
Similar articles
- Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines.
Christie PJ. Christie PJ. Mol Microbiol. 2001 Apr;40(2):294-305. doi: 10.1046/j.1365-2958.2001.02302.x. Mol Microbiol. 2001. PMID: 11309113 Free PMC article. Review. - Biological diversity of prokaryotic type IV secretion systems.
Alvarez-Martinez CE, Christie PJ. Alvarez-Martinez CE, et al. Microbiol Mol Biol Rev. 2009 Dec;73(4):775-808. doi: 10.1128/MMBR.00023-09. Microbiol Mol Biol Rev. 2009. PMID: 19946141 Free PMC article. Review. - Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells.
Christie PJ, Vogel JP. Christie PJ, et al. Trends Microbiol. 2000 Aug;8(8):354-60. doi: 10.1016/s0966-842x(00)01792-3. Trends Microbiol. 2000. PMID: 10920394 Free PMC article. Review. - The versatile bacterial type IV secretion systems.
Cascales E, Christie PJ. Cascales E, et al. Nat Rev Microbiol. 2003 Nov;1(2):137-49. doi: 10.1038/nrmicro753. Nat Rev Microbiol. 2003. PMID: 15035043 Free PMC article. Review. - Structural and dynamic properties of bacterial type IV secretion systems (review).
Christie PJ, Cascales E. Christie PJ, et al. Mol Membr Biol. 2005 Jan-Apr;22(1-2):51-61. doi: 10.1080/09687860500063316. Mol Membr Biol. 2005. PMID: 16092524 Free PMC article. Review.
Cited by
- Definition of a bacterial type IV secretion pathway for a DNA substrate.
Cascales E, Christie PJ. Cascales E, et al. Science. 2004 May 21;304(5674):1170-3. doi: 10.1126/science.1095211. Science. 2004. PMID: 15155952 Free PMC article. - Comparative genomic analysis of the pPT23A plasmid family of Pseudomonas syringae.
Zhao Y, Ma Z, Sundin GW. Zhao Y, et al. J Bacteriol. 2005 Mar;187(6):2113-26. doi: 10.1128/JB.187.6.2113-2126.2005. J Bacteriol. 2005. PMID: 15743960 Free PMC article. - The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis.
Lavander M, Ericsson SK, Bröms JE, Forsberg A. Lavander M, et al. Infect Immun. 2006 Mar;74(3):1768-76. doi: 10.1128/IAI.74.3.1768-1776.2006. Infect Immun. 2006. PMID: 16495550 Free PMC article. - A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells.
Schulein R, Guye P, Rhomberg TA, Schmid MC, Schröder G, Vergunst AC, Carena I, Dehio C. Schulein R, et al. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):856-61. doi: 10.1073/pnas.0406796102. Epub 2005 Jan 10. Proc Natl Acad Sci U S A. 2005. PMID: 15642951 Free PMC article. - Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species.
Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. Fouts DE, et al. PLoS Biol. 2005 Jan;3(1):e15. doi: 10.1371/journal.pbio.0030015. Epub 2005 Jan 4. PLoS Biol. 2005. PMID: 15660156 Free PMC article.
References
- Baron C, et al. Bacterial secrets of secretion: Euro conference on the biology of type IV secretion processes. Mol. Microbiol. 2002;43:1359–1365. - PubMed
- Salmond GPC. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 1994;32:181–200.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources