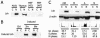Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin - PubMed (original) (raw)
Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin
Qingsheng Yan et al. Mol Cell Biol. 2003 Dec.
Abstract
The eukaryotic genome is packaged into distinct domains of transcriptionally active euchromatin and silent heterochromatin. A hallmark of mammalian heterochromatin is CpG methylation. Lsh, a member of the SNF2 family, is a major regulator of DNA methylation in mice and thus crucial for normal heterochromatin formation. In order to define the molecular function of Lsh, we examined its cellular localization and its association with chromatin. Our studies demonstrate that Lsh is an exclusively nuclear protein, and we define a nuclear localization domain within the N-terminal portion of Lsh. Lsh strongly associates with chromatin and requires the internal and C-terminal regions for this interaction. Lsh accumulates at pericentromeric heterochromatin, suggesting a direct role for Lsh in the methylation of centromeric DNA sequences and the formation of heterochromatin. In search of a signal that is responsible for Lsh recruitment to pericentromeric heterochromatin, we found that histone tail modifications were critical. Prolonged treatment with histone deacetylase inhibitors has been reported to disrupt higher-order heterochromatin organization, and this was accompanied by dissociation of Lsh from pericentromeric heterochromatin. These results are consistent with a model in which Lsh is recruited by intact heterochromatin structure and then assists in maintaining heterochromatin organization by establishing CpG methylation patterns.
Figures
FIG. 1.
Nuclear localization of Lsh. (A) Lsh is localized in nuclear extracts. For analysis of Lsh expression the pre-B-cell line PD31, fetal thymus and embryonal fibroblasts (MEFs) were examined by Western analysis and probed with a specific antibody raised against the C-terminal portion of Lsh. Lanes: C, cytoplasmic extract; N, nuclear extract (equal loading of protein extracts). (B) Lsh is localized in overexpressed nuclear extracts. Lsh was induced in 3T3 cells in the GeneSwitch system and examined by Western analysis. Lanes: C, cytoplasmic extract; N, nuclear extract (equal loading of protein extracts). (C) Lsh is preferentially expressed during S phase. The T-cell line EL4 was synchronized with mimosine (M), aphidicolin (A), or nocodazole (N) or left untreated (Ctrl) and then examined by Western analysis for the presence of Lsh or β-actin. Lanes: C, cytoplasmic extract; N, nuclear extract (extracts were prepared from equal number of cells). The lower panel shows the result of the cell cycle analysis and the distribution of cells at different stages of the cell cycle.
FIG. 2.
Nuclear localization domain of Lsh. (A) Construction map of Lsh deletional mutants. (B) Expression of deletion mutants. Nuclear extracts were examined 24 h after transfection by Western analysis with specific antiserum raised against the GFP peptide (equal loading of protein extracts). (C) The nuclear localization domain is N terminal. At 24 h after transfection with indicated plasmids, 3T3 cells were examined under a fluorescence microscope.
FIG. 3.
Lsh is associated with chromatin. (A) Lsh is present in the Triton X-resistant fraction. After Lsh induction in 3T3 cells that were transfected with the Lsh expression vector (Lsh) or transfected with the empty control vector (Ctrl) cells were extracted with Triton X-100, and the resistant and soluble fractions were examined by Western analysis for the presence of Lsh with antibodies against the C-terminal portion of Lsh (Lsh), the Flag epitope (Flag), Brg-1, MeCP2, and PCNA (extracts were prepared from equal number of cells). (B) Kinetics of Lsh expression. At different time points after Lsh induction 3T3 cells were extracted with Triton X, and the different fractions were examined by Western analysis for the presence of Lsh, PCNA, or Vimentin (extracts were prepared from equal number of cells). (C) Association with Triton X-resistant fraction is dependent on cell cycle. 3T3 cells at different stages of confluency were extracted with Triton X, and the different fractions were examined by Western analysis for the presence of Lsh or PCNA and Vimentin serving as controls (extracts were prepared from equal number of cells).(D) The C-terminal and internal domain are required for Triton X resistance. After transfection with the indicated deletion mutants, Triton X-resistant (lanes R) and soluble (Triton flush out [lanes F]) fractions were examined by Western analysis for the presence of GFP, Vimentin, and PCNA (extracts were prepared from equal number of cells).
FIG. 4.
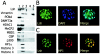
Lsh localizes to pericentromeric heterochromatin. (A) Lsh predominantly associates with chromatin. 3T3 cells were extracted with Triton X, and different fractions examined by Western analysis with the indicated antibodies: lane 1, flushed fraction; lane 2, Triton X-resistant fraction after DNase digestion; lane 3, ammonium sulfate wash; lane 4, 2 M NaCl wash; lane 5, solubilized pellet. (B) Lsh colocalizes with DAPI. At 24 h after transfection, GFP-tagged Lsh was examined by fluorescence microscopy. (C) Lsh colocalizes with HP1α. At 24 h after transfection, GFP-tagged Lsh was immunostained for detection of HP1α.
FIG. 5.
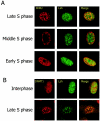
Lsh colocalizes with late replication foci. (A) Lsh colocalizes with late replication foci. Lsh was induced in 3T3 cells and immunostained with anti-Flag tag antibody and with anti-BrdU antibody for detection of incorporated BrdU. (B) Lsh colocalizes with Dnmt1. Lsh-induced 3T3 cells were immunostained with anti-Flag tag antibody and immunostained for detection of Dnmt1.
FIG. 6.
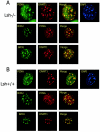
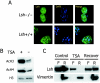
Lsh is not required for targeting of Dnmt1 to replication foci. Lsh−/− (A) or Lsh+/+ (B) MEFs were immunostained for detection of PCNA, Dnmt1, and incorporated BrdU.
FIG. 7.
Lsh dissociates from chromatin after TSA treatment. (A) Lsh associates with DAPI in hypomethylated MEFs. Lsh wild-type and Lsh−/− MEFs were transfected with a GFP-tagged Lsh expression vector and examined after 24 h. (B) TSA induces hyperacteylation. 3T3 cells were treated in the presence of 75 ng of TSA/ml, and extracts were examined by Western analysis with antibodies raised against histone 3 (H3), acetylated H3 (AcH3), or acetylated H4 (AcH4; equal loading of protein extracts). (C) Lsh dissociates from chromatin after TSA treatment and reassociates after recovery. After TSA treatment or after an additional 24 h of recovery, 3T3 cells were extracted with Triton X, and the different fractions were examined by Western analysis for the presence of induced Lsh; Vimentin served as a control. Triton X-resistant (lanes R) and soluble (Triton flush out [lanes F]) (extracts were prepared from equal number of cells) fractions are shown.
FIG. 8.
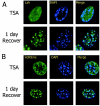
Lsh requires higher-order heterochromatin structure for localization. (A) Lsh dissociates from DAPI after TSA treatment and reassociates after recovery. After TSA treatment of 3T3 cells, Lsh was induced and cells were immunostained with anti-Flag antibody. (B) Dissociation of branched anti-H3-K9me from DAPI after TSA treatment and reassociation after recovery. The cells were also immunostained with the antibody raised against the branched H3-K9 methylated peptide.
Similar articles
- Lsh Is Essential for Maintaining Global DNA Methylation Levels in Amphibia and Fish and Interacts Directly with Dnmt1.
Dunican DS, Pennings S, Meehan RR. Dunican DS, et al. Biomed Res Int. 2015;2015:740637. doi: 10.1155/2015/740637. Epub 2015 Sep 28. Biomed Res Int. 2015. PMID: 26491684 Free PMC article. - A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association.
Han M, Li J, Cao Y, Huang Y, Li W, Zhu H, Zhao Q, Han JJ, Wu Q, Li J, Feng J, Wong J. Han M, et al. Nucleic Acids Res. 2020 Dec 2;48(21):12116-12134. doi: 10.1093/nar/gkaa1003. Nucleic Acids Res. 2020. PMID: 33170271 Free PMC article. - Lsh, a modulator of CpG methylation, is crucial for normal histone methylation.
Yan Q, Huang J, Fan T, Zhu H, Muegge K. Yan Q, et al. EMBO J. 2003 Oct 1;22(19):5154-62. doi: 10.1093/emboj/cdg493. EMBO J. 2003. PMID: 14517253 Free PMC article. - Lsh, a guardian of heterochromatin at repeat elements.
Muegge K. Muegge K. Biochem Cell Biol. 2005 Aug;83(4):548-54. doi: 10.1139/o05-119. Biochem Cell Biol. 2005. PMID: 16094458 Review. - Switching between Epigenetic States at Pericentromeric Heterochromatin.
Déjardin J. Déjardin J. Trends Genet. 2015 Nov;31(11):661-672. doi: 10.1016/j.tig.2015.09.003. Epub 2015 Sep 29. Trends Genet. 2015. PMID: 26431676 Review.
Cited by
- The C-terminal 4CXXC-type zinc finger domain of CDCA7 recognizes hemimethylated DNA and modulates activities of chromatin remodeling enzyme HELLS.
Shinkai A, Hashimoto H, Shimura C, Fujimoto H, Fukuda K, Horikoshi N, Okano M, Niwa H, Debler EW, Kurumizaka H, Shinkai Y. Shinkai A, et al. Nucleic Acids Res. 2024 Sep 23;52(17):10194-10219. doi: 10.1093/nar/gkae677. Nucleic Acids Res. 2024. PMID: 39142653 Free PMC article. - Chromatin remodeling of histone H3 variants by DDM1 underlies epigenetic inheritance of DNA methylation.
Lee SC, Adams DW, Ipsaro JJ, Cahn J, Lynn J, Kim HS, Berube B, Major V, Calarco JP, LeBlanc C, Bhattacharjee S, Ramu U, Grimanelli D, Jacob Y, Voigt P, Joshua-Tor L, Martienssen RA. Lee SC, et al. Cell. 2023 Sep 14;186(19):4100-4116.e15. doi: 10.1016/j.cell.2023.08.001. Epub 2023 Aug 28. Cell. 2023. PMID: 37643610 Free PMC article. - Chromatin remodeling of histone H3 variants underlies epigenetic inheritance of DNA methylation.
Lee SC, Adams DW, Ipsaro JJ, Cahn J, Lynn J, Kim HS, Berube B, Major V, Calarco JP, LeBlanc C, Bhattacharjee S, Ramu U, Grimanelli D, Jacob Y, Voigt P, Joshua-Tor L, Martienssen RA. Lee SC, et al. bioRxiv [Preprint]. 2023 Aug 2:2023.07.11.548598. doi: 10.1101/2023.07.11.548598. bioRxiv. 2023. PMID: 37503143 Free PMC article. Updated. Preprint. - Arabidopsis thaliana: a powerful model organism to explore histone modifications and their upstream regulations.
Yu Y, Wang S, Wang Z, Gao R, Lee J. Yu Y, et al. Epigenetics. 2023 Dec;18(1):2211362. doi: 10.1080/15592294.2023.2211362. Epigenetics. 2023. PMID: 37196184 Free PMC article. Review. - Seminars in cell and development biology on histone variants remodelers of H2A variants associated with heterochromatin.
Berger F, Muegge K, Richards EJ. Berger F, et al. Semin Cell Dev Biol. 2023 Feb 15;135:93-101. doi: 10.1016/j.semcdb.2022.02.026. Epub 2022 Mar 4. Semin Cell Dev Biol. 2023. PMID: 35249811 Free PMC article. Review.
References
- Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. - PubMed
- Bachman, K. E., B. H. Park, I. Rhee, H. Rajagopalan, J. G. Herman, S. B. Baylin, K. W. Kinzler, and B. Vogelstein. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell 3:89-95. - PubMed
- Becker, P. B., and W. Hörz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. - PubMed
- Bérubé, N. G., C. A. Smeenk, and D. J. Picketts. 2000. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum. Mol. Genet. 9:539-547. - PubMed
- Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases