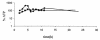Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach - PubMed (original) (raw)
Comparative Study
Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach
Christian Kohler et al. J Bacteriol. 2003 Dec.
Abstract
The high-resolution two-dimensional (2D) protein gel electrophoresis technique combined with matrix-assisted laser desorption ionization-time of flight mass spectrometry was used for identification of proteins whose levels were changed by a mutation in hemB. Cytoplasmic protein extracts obtained from the mutant and the wild type (strain COL) at different stages of growth in tryptone soya broth (exponential, transitional, and stationary growth phases) were separated on 2D protein gels. Comparison of the 2D patterns of the protein extracts of the two strains revealed major differences. Because the electron transport chain of the mutant is interrupted due to the deficiency of heme, this organism should be unable to use oxygen or nitrate as a terminal electron acceptor. Consistent with this hypothesis, proteins involved in the glycolytic pathway and related pathways (glyceraldehyde-3-phosphate dehydrogenase, enolase, and phosphoglycerate kinase) and in fermentation pathways (lactate dehydrogenase, alcohol dehydrogenase, and pyruvate formate lyase) were induced in exponentially growing cells of the mutant. These results strongly indicate that the hemB mutant generates ATP from glucose or fructose only by substrate phosphorylation. Analyses of the fermentation reactions showed that the main product was lactate. Although pyruvate formate lyase (Pfl) and pyruvate dehydrogenase were present, neither ethanol nor acetate was detected in significant amounts. Presumably, Pfl was not activated in the presence of oxygen, and pyruvate dehydrogenase might have very low activity. Transcriptional analysis of citB, encoding the aconitase, revealed that the activity of the citrate cycle enzymes was down-regulated in the hemB mutant. The arginine deiminase pathway was also induced, and it could provide ATP as well. Furthermore, the amounts of most of the extracellular virulence factors were significantly reduced by a mutation in hemB, which is consistent with previous reports.
Figures
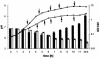
FIG. 1.
Growth (lines) and pH value (bars) of S. aureus COL (▪ and solid bars) and its isogenic hemB mutant IA48 (□ and open bars) in TSB. The times of sampling for transcriptional analysis are indicated by arrowheads on the growth curves. OD 540, optical density at 540 nm.
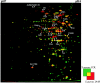
FIG. 2.
Dual-channel images of 2D gels of cytoplasmic proteins produced with the Delta2D software (Decodon GmbH), showing the differences in the protein patterns of the wild-type S. aureus strain COL (green) and the hemB mutant IA48 (red). The proteins in 100 μg of a crude cell extract from cells grown in TSB at the stationary phase were separated by preparative 2D polyacrylamide gel electrophoresis. The proteins were stained with silver nitrate. The protein spots identified are indicated by arrows. Proteins induced by a mutation in hemB are red.
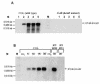
FIG. 3.
Northern blot analyses of genes whose transcription was influenced by a mutation in hemB. RNA was isolated from S. aureus COL (wild type) and its isogenic hemB mutant IA48 grown in TSB at 37°C during growth (the sampling times are indicated by arrowheads in the growth curves in Fig. 1). The membranes were hybridized with digoxigenin-labeled RNA probes for genes involved in glycolysis (A), fermentation (B), and the TCA cycle (C).
FIG. 4.
Glucose, lactate, and acetate concentrations in the supernatants of a wild-type culture (S. aureus COL) (♦) and a hemB mutant IA48 culture (▪) during growth in TSB.
FIG. 5.
Relative ATP levels in cells of S. aureus COL (♦) and its isogenic hemB mutant (▪) grown under standard conditions in TSB. The ATP concentration in the cells was determined with a Lyciferase assay kit (Roche). The light signals were detected with a Lumi-Imager and were quantified with the LumiAnalyst program from Boehringer Mannheim. The ATP level in the wild type at an optical density of 0.5 was defined as 100%.
FIG. 6.
MALDI-TOF MS analyses of Pfl. (A) Sectors of 2D gels containing the region where Pfl is located. Protein extracts of the S. aureus hemB mutant and the wild type were isolated under anaerobic (anaerob) and aerobic (aerob) conditions. The proteins were stained with silver nitrate. (B) Amino acid sequence of the pyruvate formate lyase of S. aureus. The Gly residue at position 724 that is probably radicalized is indicated by a dot. C-terminal peptide fragments 735-743 and 722-731 obtained after trypsin digestion of Pfl are indicated by boldface type. The theoretical molecular weights (monoisotopic, [MH+]) for peptides 735-743 and 722-731 are 1,086.566 and 1,096.591, respectively. (C) Sector of the MALDI-TOF MS spectrum of the 85-kDa Pfl spot covering the region for peptide 735-743 and peptide 722-731.(D) Sector of the MALDI-TOF MS spectrum of the 82-kDa Pfl spot covering the region for peptide 735-743 and peptide 722-731.
FIG. 7.
Dual-channel images of 2D gels of extracellular proteins produced with the Delta2D software (Decodon GmbH), showing the differences in the protein patterns of the wild-type S. aureus strain COL (green) and the hemB mutant IA48 (red). The proteins in 100 μg of the supernatant from cells in the stationary growth phase were separated by preparative 2D polyacrylamide gel electrophoresis. The proteins were stained with silver nitrate. Proteins induced by a mutation in hemB are red, and proteins present only in the wild-type strain are green. The following proteins that were identified are indicated by arrows: glycerophosphoryl diester phosphodiesterase (GlpQ), α-hemolysin (Hla), β-hemolysin (Hlb), immunodominant antigen A (IsaA), leucocidine F (LukF), leucocidine M (LukM), thermonuclease (Nuc), 1-phosphatidylinositol phosphodiesterase (Plc), enterotoxin B (Seb), serinproteases (SplB and SplF), and a hypothetical protein (YfnI) (38).
FIG. 8.
Northern blot analyses of RNAIII. (A) RNA was isolated from S. aureus COL (wild type) and from its isogenic hemB mutant IA48 grown in TSB at 37°C during the growth phase (the sampling times are indicated by arrowheads in Fig. 1). (B) At the stationary phase (optical density at 540 nm, 2) 20-ml portions of the hemB mutant culture were supplemented with 20-ml portions of supernatants from wild-type S. aureus strain COL, S. aureus RN6390, and the agr mutant RN6911 (all at an optical density at 540 nm of 6.5). RNA was isolated from the original culture of the mutant at zero time (co1), 30 min later (co2), and at the times indicated after addition of the supernatants to the mutant culture. The membranes were hybridized with a digoxigenin-labeled RNA probe for RNAIII.
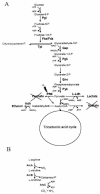
FIG. 9.
Carbohydrate (A) and arginine (B) metabolic pathways in the hemB mutant under aerobic conditions. The following enzymes were found at higher levels in the hemB mutant: phosphoglucomutase (Pgi), fructose bisphosphate aldolase (Fba/Fda), glyceraldehyde dehydrogenase (Gap), phosphoglycerate kinase (Pgk), enolase (Eno), pyruvate kinase (Pyk),
l
-lactate dehydrogenase (L-Ldh), pyruvate formate lyase (PflB), alcohol dehydrogenase (Adh), arginine deiminase (ArcA), ornithine carbamoyltransferase (ArcB), and carbamate kinase (ArcC).
Similar articles
- A defect in menadione biosynthesis induces global changes in gene expression in Staphylococcus aureus.
Kohler C, von Eiff C, Liebeke M, McNamara PJ, Lalk M, Proctor RA, Hecker M, Engelmann S. Kohler C, et al. J Bacteriol. 2008 Oct;190(19):6351-64. doi: 10.1128/JB.00505-08. Epub 2008 Aug 1. J Bacteriol. 2008. PMID: 18676673 Free PMC article. - Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus.
Weber H, Engelmann S, Becher D, Hecker M. Weber H, et al. Mol Microbiol. 2004 Apr;52(1):133-40. doi: 10.1111/j.1365-2958.2004.03971.x. Mol Microbiol. 2004. PMID: 15049816 - Small colony variants of Staphylococcus aureus reveal distinct protein profiles.
Kriegeskorte A, König S, Sander G, Pirkl A, Mahabir E, Proctor RA, von Eiff C, Peters G, Becker K. Kriegeskorte A, et al. Proteomics. 2011 Jun;11(12):2476-90. doi: 10.1002/pmic.201000796. Epub 2011 May 18. Proteomics. 2011. PMID: 21595038 - Proteome analyses of Staphylococcus aureus in growing and non-growing cells: a physiological approach.
Kohler C, Wolff S, Albrecht D, Fuchs S, Becher D, Büttner K, Engelmann S, Hecker M. Kohler C, et al. Int J Med Microbiol. 2005 Dec;295(8):547-65. doi: 10.1016/j.ijmm.2005.08.002. Epub 2005 Oct 25. Int J Med Microbiol. 2005. PMID: 16325551 - X-ray studies of glycolytic enzymes.
Blake CC. Blake CC. Essays Biochem. 1975;11:37-79. Essays Biochem. 1975. PMID: 174910 Review. No abstract available.
Cited by
- Staphylococcus aureus ArcR controls expression of the arginine deiminase operon.
Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. Makhlin J, et al. J Bacteriol. 2007 Aug;189(16):5976-86. doi: 10.1128/JB.00592-07. Epub 2007 Jun 8. J Bacteriol. 2007. PMID: 17557828 Free PMC article. - Hinokitiol potentiates antimicrobial activity of bismuth drugs: a combination therapy for overcoming antimicrobial resistance.
Ip TK, Wang Y, Wang S, Pu K, Wang R, Han B, Gao P, Xie Y, Kao RY, Ho PL, Li H, Sun H. Ip TK, et al. RSC Med Chem. 2025 Jan 31. doi: 10.1039/d4md00860j. Online ahead of print. RSC Med Chem. 2025. PMID: 40027343 Free PMC article. - Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB.
Moisan H, Brouillette E, Jacob CL, Langlois-Bégin P, Michaud S, Malouin F. Moisan H, et al. J Bacteriol. 2006 Jan;188(1):64-76. doi: 10.1128/JB.188.1.64-76.2006. J Bacteriol. 2006. PMID: 16352822 Free PMC article. - Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype.
von Eiff C, McNamara P, Becker K, Bates D, Lei XH, Ziman M, Bochner BR, Peters G, Proctor RA. von Eiff C, et al. J Bacteriol. 2006 Jan;188(2):687-93. doi: 10.1128/JB.188.2.687-693.2006. J Bacteriol. 2006. PMID: 16385058 Free PMC article. - Insights into the mode of action of chitosan as an antibacterial compound.
Raafat D, von Bargen K, Haas A, Sahl HG. Raafat D, et al. Appl Environ Microbiol. 2008 Jun;74(12):3764-73. doi: 10.1128/AEM.00453-08. Epub 2008 May 2. Appl Environ Microbiol. 2008. PMID: 18456858 Free PMC article.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. - PubMed
- Bernhardt, J., K. Büttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225-2240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials