La, PTB, and PAB proteins bind to the 3(') untranslated region of Norwalk virus genomic RNA - PubMed (original) (raw)
La, PTB, and PAB proteins bind to the 3(') untranslated region of Norwalk virus genomic RNA
Ana Lorena Gutiérrez-Escolano et al. Biochem Biophys Res Commun. 2003.
Abstract
Noroviruses are human enteric caliciviruses for which no cell culture is available. Consequently, the mechanisms and factors involved in their replication have been difficult to study. In an attempt to analyze the cis- and trans-acting factors that could have a role in NV replication, the 3(')-untranslated region of the genome was studied. Use of Zuker's mfold-2 software predicted that NV 3(')UTR contains a stem-loop structure of 47 nts. Proteins from HeLa cell extracts, such as La and PTB, form stable complexes with this region. The addition of a poly(A) tail (24 nts) to the 3(')UTR permits the specific binding of the poly(A) binding protein (PABP) present in HeLa cell extracts, as well as the recombinant PABP. Since La, PTB, and PABP are important trans-acting factors required for viral translation and replication, these RNA-protein interactions may play a role in NV replication or translation.
Figures
Fig. 1
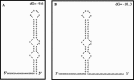
Schematic representation of the NV 3′UTR secondary structure. (A) Predicted secondary structure of the 66 nt long 3′UTR, which forms the complete 3′UTR of NV genome. (B) Predicted secondary structure of the same region plus a 24 nt long poly(A) tail. Both predictions were performed using the mfold-2 software (
).
Fig. 2
Mobility-shift analysis of the complete 3′UTR of NV and S10 extract from HeLa cell. (A) [α-32P]UTP labeled 3′UTR incubated without (lane 1) or with 5, 10, 15, and 20 μg of S10 extract from HeLa cells (lanes 2–5, respectively). (B) [α-32P]UTP labeled 3′ UTR was incubated without (lane 1) or with 5, 10, 15, and 20 μg of HeLa S10 extract (lanes 2–5, respectively) followed by RNase treatment. (C) [α-32P]UTP labeled 3′UTR RNA was incubated without (lane 1) or with 20 μg S10 extract from HeLa cells (lanes 2–5), in the absence (lanes 2) or, the presence of 0.6, 0.9, and 1.2 M KCl (lanes 3–5, respectively). (D) [α-32P]UTP labeled 3′UTR RNA was incubated without (lane 1) or with 20 μg of S10 extract from HeLa cells (2–5) in the absence (lane 2) or presence of 10- and 20-fold molar excess of homologous (lanes 3 and 4, respectively) or 20-fold molar excess of non-related heterologous competitor (lane 5). Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Mobility of complexes I and II is indicated.
Fig. 3
Mobility-shift analysis of the complete 3′UTR and the 3′UTR (A) of NV and S10 extract from HeLa cells. (A) [α-32P]UTP labeled 3′UTR (lanes 1 and 2) or 3′UTR(A) RNAs were incubated in the absence (lane 1) or presence of 20 μg of S10 extract from HeLa cells (lane 2 and 3) followed by RNAse treatment. (B) [α-32P]ATP labeled 3′UTR (lane 3) or 3′UTR(A) RNAs (lanes 1 and 2) were incubated in the absence (lane 1) or presence of 10 μg of S10 extract from HeLa cells (lanes 2 and 3) followed by RNAse treatment. Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Mobility of complexes is indicated on both sides of the figure.
Fig. 4
Specificity of RNA–protein complexes formed with the complete 3′UTR and the 3′UTR(A) of NV with S10 extract from HeLa cells. (A) [α-32P]ATP labeled 3′UTR(A) RNA was incubated with 10 μg of S10 extract from HeLa cells in the absence (lanes 1 and 2) or presence of 25-fold molar excess of unlabeled homologous (lane 3), heterologous (lane 4), and non-related heterologous competitors (lane 5). (B) Ten micrograms of S10 extract from HeLa cells was incubated in the absence (lanes 1 and 2) or in the presence of 1.5 μl of polyclonal antibodies to human PAB protein (lane 3) or an anti-GADPH antibody (lane 4) before the addition of [α-32P]ATP labeled 3′UTR(A) RNA. The antibody–RNA–protein supercomplex was further processed under the same conditions described for the RNA–protein complex. (C) [α-32P]ATP labeled 3′UTR RNA (lane 2) or 3′UTR(A) (lanes 1 and 3) was incubated in the absence (lane 1) or presence of 100 ng of recombinant PAB protein (lanes 2 and 3) followed by RNase treatment. Complex formation was assayed by electrophoresis on native polyacrylamide gels and detected by autoradiography. Free RNA was loaded on lane 1. Mobility of complexes formed with S10 HeLa cell extract and with the rPABP is indicated on the right-hand side of the figure.
Fig. 5
UV-induced cross-linking of the complete 3′UTR and the 3′UTR (A) of NV and S10 extract from HeLa cells. (A) [α-32P]ATP labeled 3′UTR (lanes 1 and 2) and 3′UTR(A) (lane 3) RNAs were UV-crosslinked without (lane 1) or with 60 μg of S10 extract from HeLa cells (lanes 2–3). (B) Forty micrograms of S10 extract from HeLa cells were preincubated 15 min with 25-fold molar excess of heterologous (lane 2), homologous (lane 3), and non-related heterologous (lane 5) RNAs prior to the addition of [α-32P]ATP labeled 3′UTR(A). Free RNA was loaded on lane 1. (C) [α-32P]UTP labeled 3′UTR RNA was cross-linked with 60 μg of S10 extract from HeLa cells (lane 1) and immunoprecipitated with anti-La (lane 3) or anti-actin antibodies (lane 2). (D) [α-32P]UTP labeled 3′UTR RNA was UV-cross-linked with 60 μg of S10 extract from HeLa cells (lane 1) or 100 and 500 ng of recombinant PTB protein (lanes 2 and 3, respectively). After UV-cross-linking, the reaction was followed by RNase treatment. Crosslinked proteins were loaded on an SDS–10% polyacrylamide gel and detected by autoradiography. The migration of the UV-crosslinked 68 kDa, rPTB, and the immunoprecipitated La proteins is indicated by an arrow.
References
- Fields N.B., Knipe D.M., Howley P.M., editors. Human Caliciviruses. Lippincott, Williams, and Wilkins (Ed.); Philadelphia, USA: 2001. pp. 841–874. (Virology, fourth ed.).
- Frankhouser R.L., Noel J.S., Monroe S.S., Ando T., Glass R.I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1998;178:1571–1578. - PubMed
- Subekti D.S., Tjaniadi P., Lsmana M., McArdle J., Iskandriati D., Budiarsa I.N., Walujo P., Suparto H.I., Winoto I., Campbell J.R., Porter K.R., Sajuthi K.D., Ansari A.A., Oyofo B.A. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 2002;66:400–406. - PubMed
- Jiang X., Wang X.M., Wang K., Estes M.K. Sequence and genomic organization of norwalk virus. Virology. 1993;195:51–61. - PubMed
- Clarke N., Lambden P.R. The molecular biology of caliciviruses. J. Gen. Virol. 1997;78:291–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources