Viral discovery and sequence recovery using DNA microarrays - PubMed (original) (raw)
doi: 10.1371/journal.pbio.0000002. Epub 2003 Nov 17.
Anatoly Urisman, Yu-Tsueng Liu, Michael Springer, Thomas G Ksiazek, Dean D Erdman, Elaine R Mardis, Matthew Hickenbotham, Vincent Magrini, James Eldred, J Phillipe Latreille, Richard K Wilson, Don Ganem, Joseph L DeRisi
Affiliations
- PMID: 14624234
- PMCID: PMC261870
- DOI: 10.1371/journal.pbio.0000002
Viral discovery and sequence recovery using DNA microarrays
David Wang et al. PLoS Biol. 2003 Nov.
Abstract
Because of the constant threat posed by emerging infectious diseases and the limitations of existing approaches used to identify new pathogens, there is a great demand for new technological methods for viral discovery. We describe herein a DNA microarray-based platform for novel virus identification and characterization. Central to this approach was a DNA microarray designed to detect a wide range of known viruses as well as novel members of existing viral families; this microarray contained the most highly conserved 70mer sequences from every fully sequenced reference viral genome in GenBank. During an outbreak of severe acute respiratory syndrome (SARS) in March 2003, hybridization to this microarray revealed the presence of a previously uncharacterized coronavirus in a viral isolate cultivated from a SARS patient. To further characterize this new virus, approximately 1 kb of the unknown virus genome was cloned by physically recovering viral sequences hybridized to individual array elements. Sequencing of these fragments confirmed that the virus was indeed a new member of the coronavirus family. This combination of array hybridization followed by direct viral sequence recovery should prove to be a general strategy for the rapid identification and characterization of novel viruses and emerging infectious disease.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
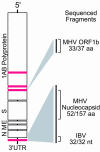
Figure 1. Prototypical Coronavirus Genome Structure
Red bars indicate physical location of virus microarray DNA elements mapped to a generic coronavirus genome. Portions of the coronavirus genome sequenced by physical recovery and PCR methods are highlighted with homologies to known coronaviruses. Abbreviations: aa, amino acid; nt, nucleotide
Figure 2. Viral DNA Recovery and Sequencing Scheme
Hybridized viral sequences were physically scraped from a DNA microarray spot, amplified, cloned, and subsequently sequenced.
Similar articles
- Design of microarray probes for virus identification and detection of emerging viruses at the genus level.
Chou CC, Lee TT, Chen CH, Hsiao HY, Lin YL, Ho MS, Yang PC, Peck K. Chou CC, et al. BMC Bioinformatics. 2006 Apr 28;7:232. doi: 10.1186/1471-2105-7-232. BMC Bioinformatics. 2006. PMID: 16643672 Free PMC article. - Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays.
Wong CW, Albert TJ, Vega VB, Norton JE, Cutler DJ, Richmond TA, Stanton LW, Liu ET, Miller LD. Wong CW, et al. Genome Res. 2004 Mar;14(3):398-405. doi: 10.1101/gr.2141004. Genome Res. 2004. PMID: 14993206 Free PMC article. - [SARS-associated coronavirus gene fragments were detected from a suspected pediatric SARS patient].
Zhu RN, Qian Y, Deng J, Zhao LQ, Wang F, Cao L, Wang TY, Chen DK, Zhang Q. Zhu RN, et al. Zhonghua Er Ke Za Zhi. 2003 Sep;41(9):641-4. Zhonghua Er Ke Za Zhi. 2003. PMID: 14733796 Chinese. - From genome to antivirals: SARS as a test tube.
Kliger Y, Levanon EY, Gerber D. Kliger Y, et al. Drug Discov Today. 2005 Mar 1;10(5):345-52. doi: 10.1016/S1359-6446(04)03320-3. Drug Discov Today. 2005. PMID: 15749283 Free PMC article. Review. - [Resequencing microarrays: a rapid tool for better identification and understanding of viral and bacterial emergence].
Berthet N. Berthet N. Bull Acad Natl Med. 2013 Dec;197(9):1669-82. Bull Acad Natl Med. 2013. PMID: 26137813 Review. French.
Cited by
- Viral pathogen discovery.
Chiu CY. Chiu CY. Curr Opin Microbiol. 2013 Aug;16(4):468-78. doi: 10.1016/j.mib.2013.05.001. Epub 2013 May 29. Curr Opin Microbiol. 2013. PMID: 23725672 Free PMC article. Review. - Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes.
Jiang H, Holtz LR, Bauer I, Franz CJ, Zhao G, Bodhidatta L, Shrestha SK, Kang G, Wang D. Jiang H, et al. Virology. 2013 Feb 5;436(1):8-14. doi: 10.1016/j.virol.2012.09.040. Epub 2012 Oct 16. Virology. 2013. PMID: 23084422 Free PMC article. - Application of next-generation sequencing to identify different pathogens.
Nafea AM, Wang Y, Wang D, Salama AM, Aziz MA, Xu S, Tong Y. Nafea AM, et al. Front Microbiol. 2024 Jan 29;14:1329330. doi: 10.3389/fmicb.2023.1329330. eCollection 2023. Front Microbiol. 2024. PMID: 38348304 Free PMC article. Review. - Optimization and clinical validation of a pathogen detection microarray.
Wong CW, Heng CL, Wan Yee L, Soh SW, Kartasasmita CB, Simoes EA, Hibberd ML, Sung WK, Miller LD. Wong CW, et al. Genome Biol. 2007;8(5):R93. doi: 10.1186/gb-2007-8-5-r93. Genome Biol. 2007. PMID: 17531104 Free PMC article. - Rapid identification of emerging pathogens: coronavirus.
Sampath R, Hofstadler SA, Blyn LB, Eshoo MW, Hall TA, Massire C, Levene HM, Hannis JC, Harrell PM, Neuman B, Buchmeier MJ, Jiang Y, Ranken R, Drader JJ, Samant V, Griffey RH, McNeil JA, Crooke ST, Ecker DJ. Sampath R, et al. Emerg Infect Dis. 2005 Mar;11(3):373-9. doi: 10.3201/eid1103.040629. Emerg Infect Dis. 2005. PMID: 15757550 Free PMC article.
References
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. - PubMed
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. - PubMed
- Jonassen CM, Jonassen TO, Grinde B. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J Gen Virol. 1998;79:715–718. - PubMed
- Kellam P. Molecular identification of novel viruses. Trends Microbiol. 1998;6:160–165. - PubMed
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous