Identifying cooperativity among transcription factors controlling the cell cycle in yeast - PubMed (original) (raw)
Identifying cooperativity among transcription factors controlling the cell cycle in yeast
Nilanjana Banerjee et al. Nucleic Acids Res. 2003.
Abstract
Transcription regulation in eukaryotes is known to occur through the coordinated action of multiple transcription factors (TFs). Recently, a few genome-wide transcription studies have begun to explore the combinatorial nature of TF interactions. We propose a novel approach that reveals how multiple TFs cooperate to regulate transcription in the yeast cell cycle. Our method integrates genome-wide gene expression data and chromatin immunoprecipitation (ChIP-chip) data to discover more biologically relevant synergistic interactions between different TFs and their target genes than previous studies. Given any pair of TFs A and B, we define a novel measure of cooperativity between the two TFs based on the expression patterns of sets of target genes of only A, only B, and both A and B. If the cooperativity measure is significant then there is reason to postulate that the presence of both TFs is needed to influence gene expression. Our results indicate that many cooperative TFs that were previously characterized experimentally indeed have high values of cooperativity measures in our analysis. In addition, we propose several novel, experimentally testable predictions of cooperative TFs that play a role in the cell cycle and other biological processes. Many of them hold interesting clues for cross talk between the cell cycle and other processes including metabolism, stress response and pseudohyphal differentiation. Finally, we have created a web tool where researchers can explore the exhaustive list of cooperative TFs and survey the graphical representation of the target genes' expression profiles. The interface includes a tool to dynamically draw a TF cooperativity network of 113 TFs with user-defined significance levels. This study is an example of how systematic combination of diverse data types along with new functional genomic approaches can provide a rigorous platform to map TF interactions more efficiently.
Figures
Figure 1
Schematic diagram of the ChIP-based approach of identifying cooperativity. The promoters of the set of genes that show binding of both TFs A (TFA) and B (TFB) show a much higher EC score (0.22) than the promoters of the sets of genes that show binding to only TF A (0.05) or TF B (0.06). Therefore, TFs A and B are potentially cooperative.
Figure 2
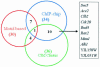
Overlapping ORFs in ChIP, motif and expression analysis. ChIP-based and expression-based approaches appear to identify more experimentally established targets of Mcm1 and Fkh2 than the motif- and expression-based approach described in Pilpel et al. (3).
Figure 3
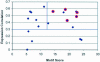
Motif scores and gene expression of ChIP-positive target genes of Mcm1 and Fkh2. The known targets (circled in red) fall in the upper right-hand corner. The genes that are close to the known target genes in expression and motif space are likely to be reliable additional targets of Mcm1 and Fkh2 compared with those genes with low correlation with core expression profile and low motif score.
Figure 4
User interface to get a TF combination’s target genes and their expression profiles.
Figure 5
User interface to dynamically generate a TF cooperativity network with user-defined significance values.
Similar articles
- Identifying cooperative transcription factors in yeast using multiple data sources.
Lai FJ, Jhu MH, Chiu CC, Huang YM, Wu WS. Lai FJ, et al. BMC Syst Biol. 2014;8 Suppl 5(Suppl 5):S2. doi: 10.1186/1752-0509-8-S5-S2. Epub 2014 Dec 12. BMC Syst Biol. 2014. PMID: 25559499 Free PMC article. - Properly defining the targets of a transcription factor significantly improves the computational identification of cooperative transcription factor pairs in yeast.
Wu WS, Lai FJ. Wu WS, et al. BMC Genomics. 2015;16 Suppl 12(Suppl 12):S10. doi: 10.1186/1471-2164-16-S12-S10. Epub 2015 Dec 9. BMC Genomics. 2015. PMID: 26679776 Free PMC article. - Identifying combinatorial regulation of transcription factors and binding motifs.
Kato M, Hata N, Banerjee N, Futcher B, Zhang MQ. Kato M, et al. Genome Biol. 2004;5(8):R56. doi: 10.1186/gb-2004-5-8-r56. Epub 2004 Jul 28. Genome Biol. 2004. PMID: 15287978 Free PMC article. - A conserved role for transcription factor sumoylation in binding-site selection.
Rosonina E. Rosonina E. Curr Genet. 2019 Dec;65(6):1307-1312. doi: 10.1007/s00294-019-00992-w. Epub 2019 May 15. Curr Genet. 2019. PMID: 31093693 Review. - Mapping yeast transcriptional networks.
Hughes TR, de Boer CG. Hughes TR, et al. Genetics. 2013 Sep;195(1):9-36. doi: 10.1534/genetics.113.153262. Genetics. 2013. PMID: 24018767 Free PMC article. Review.
Cited by
- A systematic study of HIF1A cofactors in hypoxic cancer cells.
Zhang Y, Wang S, Hu H, Li X. Zhang Y, et al. Sci Rep. 2022 Nov 8;12(1):18962. doi: 10.1038/s41598-022-23060-9. Sci Rep. 2022. PMID: 36347941 Free PMC article. - TFSyntax: a database of transcription factors binding syntax in mammalian genomes.
Zhao Y. Zhao Y. Nucleic Acids Res. 2023 Jan 6;51(D1):D306-D314. doi: 10.1093/nar/gkac849. Nucleic Acids Res. 2023. PMID: 36200824 Free PMC article. - Anthocyanins accumulation analysis of correlated genes by metabolome and transcriptome in green and purple peppers (Capsicum annuum).
Meng Y, Zhang H, Fan Y, Yan L. Meng Y, et al. BMC Plant Biol. 2022 Jul 22;22(1):358. doi: 10.1186/s12870-022-03746-y. BMC Plant Biol. 2022. PMID: 35869427 Free PMC article. - Bayesian regularization for a nonstationary Gaussian linear mixed effects model.
Gecili E, Sivaganesan S, Asar O, Clancy JP, Ziady A, Szczesniak RD. Gecili E, et al. Stat Med. 2022 Feb 20;41(4):681-697. doi: 10.1002/sim.9279. Epub 2021 Dec 12. Stat Med. 2022. PMID: 34897771 Free PMC article. - Reciprocal stabilization of transcription factor binding integrates two signaling pathways to regulate fission yeast fbp1 transcription.
Koda W, Senmatsu S, Abe T, Hoffman CS, Hirota K. Koda W, et al. Nucleic Acids Res. 2021 Sep 27;49(17):9809-9820. doi: 10.1093/nar/gkab758. Nucleic Acids Res. 2021. PMID: 34486060 Free PMC article.
References
- Wagner A. (1999) Genes regulated cooperatively by one or more transcription factors and their identification in whole eukaryotic genomes. Bioinformatics, 15, 776–784. - PubMed
- Pilpel Y., Sudarsanam,P. and Church,G. (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nature Genet., 29, 153–159. - PubMed
- Lee T.I., Rinaldi,N.J., Robert,F., Odom,D.T., Bar-Joseph,Z., Gerber,G.K., Hannett,N.M., Harbison,C.T., Thompson,C.M., Simon,I. et al. (2002) Transcriptional regulatory networks in S. cerevisiae. Science, 298, 799–804. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous