Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time - PubMed (original) (raw)
. 2003 Dec;15(12):2856-65.
doi: 10.1105/tpc.016345. Epub 2003 Nov 20.
Affiliations
- PMID: 14630968
- PMCID: PMC282816
- DOI: 10.1105/tpc.016345
Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time
Shinobu Takada et al. Plant Cell. 2003 Dec.
Abstract
The flowering time of plants is tightly regulated by both promotive and repressive factors. Molecular genetic studies using Arabidopsis have identified several epigenetic repressors that regulate flowering time. Terminal flower2, (TFL2), which encodes a homolog of heterochromatin protein1 represses flowering locus T (FT) expression, which is induced by the activator constans (CO) in response to the long-day signal. Here, we show that TFL2, CO, and FT are expressed together in leaf vascular tissues and that TFL2 represses FT expression continuously throughout development. Mutations in TFL2 derepress FT expression within the vascular tissues of leaves, resulting in daylength-independent early flowering. TFL2 can reduce FT expression even when CO is overexpressed. However, FT expression reaches a level sufficient for floral induction even in the presence of TFL2, suggesting that TFL2 does not maintain FT in a silent state or inhibit it completely; rather, it counteracts the effect of CO on FT activation.
Figures
Figure 1.
Whole-Mount Expression Analyses of CO, FT, and TFL2. (A) Constructs used in expression analyses (see Methods). Yellow boxes indicate open reading frames. Blue boxes indicate the uidA open reading frame (GUS gene). ATG and STOP indicate what actually function, so that only gTFL2:GUS encodes a fusion protein. nost, the nopaline synthase terminator. (B) to (Q) GUS expression patterns of gCO:GUS ([B], [F], [J], and [N]), gTFL2:GUS ([C], [G], [K], and [O]), and pFT:GUS ([D], [H], [L], and [P]) in ecotype Columbia (Col) and pFT:GUS in tfl2-2 ([E], [I], [M], and [Q]) in whole-mount staining of 6-day-old seedlings ([B] to [E]), 12-day-old seedlings ([F] to [I]), the first true leaves of 6-day-old seedlings ([J] to [M]), and the first true leaves of 8-day-old seedlings ([N] to [Q]). (R) to (T) GUS expression in short-day conditions of 12-day-old gCO:GUS (R), 6-day-old pFT:GUS (S), and 12-day-old pFT:GUS (T) plants. The inset in (S) shows a higher magnification of the first true leaf. (U) In situ hybridization against FT mRNA in the cotyledon of tfl2-2 (longitudinal section). The arrowhead indicates GUS expression in the primary vein in (M). Bars = 1 mm in (B) for (B) to (E), 1 mm in (F) for (F) to (H), 1 mm in (I), 0.1 mm in (J) for (J) to (M), 0.1 mm in (N) for (N) to (Q), 1 mm in (R) to (T), and 0.1 mm in (U).
Figure 2.
Histological Analyses of GUS Expression Patterns. (A) to (D) Longitudinal sections through 8-day-old seedlings of gCO:GUS (A), gTFL2:GUS (B), and pFT:GUS (C) in Col and pFT:GUS in tfl2-2 (D). (E) to (H) Transverse sections through leaves of gCO:GUS (E), gTFL2:GUS (F), and pFT:GUS (G) in Col and pFT:GUS in tfl2-2 (H). Asterisks indicates the SAM. P, phloem; X, xylem. Bars = 0.1 mm in (A) for (A) to (D), 10 μm in (E) for (E) to (H), and 10 μm for the inset in (G).
Figure 3.
tfl2 Affects Both the _CO_-Dependent and _CO_-Independent Expression of FT. (A) Quantitative GUS expression analysis of pFT:GUS in Col (solid line) and tfl2-2 (dashed line). GUS activity is shown as the mean ±
se
of 4-methylumbelliferyl glucuronide·min−1·μg−1 protein of three independent experiments. (B) to (D) Real-time quantitative RT-PCR analysis of FT ([B] and [C]) and CO (D) expression. The same RNA extract was used for the experiments shown in (B) and (D). These data are normalized to the amount of ACT2 (set as 100%) and are means ±
se
of three independent experiments. (B) FT expression levels in 6-day-old Col, tfl2-2, co-101, and co-101 tfl2-2 seedlings. (C) FT expression over time in Col, tfl2-2, co-101, and co-101 tfl2-2 seedlings. (D) CO expression levels in 6-day-old Col and tfl2-2 seedlings. (E) Flowering time measured as the mean number (±
se
) of rosette leaves at flowering in Col, co-101, tfl2-2, and co-101 tfl2-2 (n = 27, 7, 27, and 9, respectively). All plants were grown under long-day conditions. Asterisks indicate data that do not show statistically significant differences.
Figure 4.
pFT:GUS Expression Requires CO Activity. pFT:GUS expression is shown in 12-day-old seedlings of wild-type Col (A), co-101 (B), and co-101 tfl2-2 (C) (roots were cut off for genotyping) and in leaves of wild-type Col (D), co-101 (E), and co-101 tfl2-2 (F). Bars = 1 mm.
Figure 5.
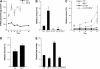
Roles of TFL2 during FT Repression in the CO Overexpressor. (A) Real-time quantitative RT-PCR analysis of FT (closed bars) and CO (open bars) expression in 12-day-old seedlings of Col, tfl2-2, 35S:CO, and 35S:CO tfl2-2. The data are normalized to the amount of ACT2 (set as 100%) and are means ±
se
of three independent experiments. There was no statistically significant difference among genotypes marked with the same symbols. (B) Flowering time measured as the mean number (±
se
) of rosette leaves at flowering in Col, tfl2-2, 35S:CO, and 35S:CO tfl2-2 (n = 27, 27, 43, and 22, respectively). (C) and (D) Whole-mount analysis of pFT:GUS expression in 35S:CO (C) and 35S:CO tfl2-2 (D). Bars = 1 mm.
Similar articles
- Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes.
Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. Kotake T, et al. Plant Cell Physiol. 2003 Jun;44(6):555-64. doi: 10.1093/pcp/pcg091. Plant Cell Physiol. 2003. PMID: 12826620 - Enabling photoperiodic control of flowering by timely chromatin silencing of the florigen gene.
He Y. He Y. Nucleus. 2015;6(3):179-82. doi: 10.1080/19491034.2015.1038000. Epub 2015 May 7. Nucleus. 2015. PMID: 25950625 Free PMC article. - The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering.
Castillejo C, Pelaz S. Castillejo C, et al. Curr Biol. 2008 Sep 9;18(17):1338-43. doi: 10.1016/j.cub.2008.07.075. Epub 2008 Aug 21. Curr Biol. 2008. PMID: 18718758 - Just say no: floral repressors help Arabidopsis bide the time.
Yant L, Mathieu J, Schmid M. Yant L, et al. Curr Opin Plant Biol. 2009 Oct;12(5):580-6. doi: 10.1016/j.pbi.2009.07.006. Epub 2009 Aug 18. Curr Opin Plant Biol. 2009. PMID: 19695946 Review. - Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration.
Shim JS, Kubota A, Imaizumi T. Shim JS, et al. Plant Physiol. 2017 Jan;173(1):5-15. doi: 10.1104/pp.16.01327. Epub 2016 Sep 29. Plant Physiol. 2017. PMID: 27688622 Free PMC article. Review.
Cited by
- FLOWERING LOCUS T has higher protein mobility than TWIN SISTER OF FT.
Jin S, Jung HS, Chung KS, Lee JH, Ahn JH. Jin S, et al. J Exp Bot. 2015 Oct;66(20):6109-17. doi: 10.1093/jxb/erv326. Epub 2015 Jul 2. J Exp Bot. 2015. PMID: 26139827 Free PMC article. - SHAGGY-like kinase 12 regulates flowering through mediating CONSTANS stability in Arabidopsis.
Chen Y, Song S, Gan Y, Jiang L, Yu H, Shen L. Chen Y, et al. Sci Adv. 2020 Jun 12;6(24):eaaw0413. doi: 10.1126/sciadv.aaw0413. eCollection 2020 Jun. Sci Adv. 2020. PMID: 32582842 Free PMC article. - Fruit load modulates flowering-related gene expression in buds of alternate-bearing 'Moncada' mandarin.
Muñoz-Fambuena N, Mesejo C, González-Mas MC, Primo-Millo E, Agustí M, Iglesias DJ. Muñoz-Fambuena N, et al. Ann Bot. 2012 Nov;110(6):1109-18. doi: 10.1093/aob/mcs190. Epub 2012 Aug 21. Ann Bot. 2012. PMID: 22915579 Free PMC article. - GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance.
Sun H, Jia Z, Cao D, Jiang B, Wu C, Hou W, Liu Y, Fei Z, Zhao D, Han T. Sun H, et al. PLoS One. 2011;6(12):e29238. doi: 10.1371/journal.pone.0029238. Epub 2011 Dec 14. PLoS One. 2011. PMID: 22195028 Free PMC article. - Poplar FT2 shortens the juvenile phase and promotes seasonal flowering.
Hsu CY, Liu Y, Luthe DS, Yuceer C. Hsu CY, et al. Plant Cell. 2006 Aug;18(8):1846-61. doi: 10.1105/tpc.106.041038. Epub 2006 Jul 14. Plant Cell. 2006. PMID: 16844908 Free PMC article.
References
- Ahmad, K., and Henikoff, S. (2002). Epigenetic consequences of nucleosome dynamics. Cell 111, 281–284. - PubMed
- Araki, T. (2001). Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4, 63–68. - PubMed
- Blazquez, M.A., Ahn, J.H., and Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet 33, 168–171. - PubMed
- Blazquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. - PubMed
- Colasanti, J., and Sundaresan, V. (2000). ‘Florigen’ enters the molecular age: Long-distance signals that cause plants to flower. Trends Biochem. Sci. 25, 236–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases