Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes - PubMed (original) (raw)
Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of Radical SAM enzymes
Gunhild Layer et al. EMBO J. 2003.
Abstract
'Radical SAM' enzymes generate catalytic radicals by combining a 4Fe-4S cluster and S-adenosylmethionine (SAM) in close proximity. We present the first crystal structure of a Radical SAM enzyme, that of HemN, the Escherichia coli oxygen-independent coproporphyrinogen III oxidase, at 2.07 A resolution. HemN catalyzes the essential conversion of coproporphyrinogen III to protoporphyrinogen IX during heme biosynthesis. HemN binds a 4Fe-4S cluster through three cysteine residues conserved in all Radical SAM enzymes. A juxtaposed SAM coordinates the fourth Fe ion through its amide nitrogen and carboxylate oxygen. The SAM sulfonium sulfur is near both the Fe (3.5 A) and a neighboring sulfur of the cluster (3.6 A), allowing single electron transfer from the 4Fe-4S cluster to the SAM sulfonium. SAM is cleaved yielding a highly oxidizing 5'-deoxyadenosyl radical. HemN, strikingly, binds a second SAM immediately adjacent to the first. It may thus successively catalyze two propionate decarboxylations. The structure of HemN reveals the cofactor geometry required for Radical SAM catalysis and sets the stage for the development of inhibitors with antibacterial function due to the uniquely bacterial occurrence of the enzyme.
Figures
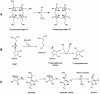
Fig. 1. Schematic representation of the enzymatic reaction of HemN. (A) HemN oxidatively decarboxylates coproporphyrinogen III to protoporphyrinogen IX by converting the propionate side chains of rings A and B to the corresponding vinyl groups. (B) The first reaction step common to HemN and all Radical SAM enzymes: a reduced 4Fe–4S cluster transfers an electron to the sulfonium of _S_-adenosylmethionine (SAM). The C5′–S+ bond of SAM is cleaved, producing methionine and a highly oxidizing 5′-deoxyadenosyl radical. The radical abstracts a hydrogen atom from a substrate RH (the substrate may itself be an enzyme), creating the corresponding substrate radical (R·). (C) In the reaction catalyzed by HemN, the 5′-deoxyadenosyl radical abstracts a hydrogen atom from the β-C atom of the substrate propionate side chain. CO2 is eliminated, and a single electron transfer to an electron acceptor gives rise to the vinyl group of the reaction product.
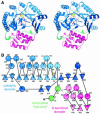
Fig. 2. Structure of HemN. (A) A ribbons-type and (B) a schematic representation of the secondary structure elements. HemN consists of two distinct domains (shades of blue and red) as well as an elongated N-terminal region termed a trip-wire (green). The catalytic domain is built around a 12-stranded, largely parallel β-sheet. At its core, the N-terminal region bears a three-quarter barrel, a (βα)6 variation of the (βα)8 TIM barrel. This core binds all cofactors, a 4Fe–4S cluster and two SAM molecules. The N-terminal trip-wire and the C-terminal domain probably participate in substrate binding. A CxxxCxxC motif, conserved in all Radical SAM proteins, is located in a loop following the first β-strand of the central barrel. The three cysteines (small yellow circles) bind three of the Fe ions of the cluster.
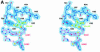
Fig. 3. Detailed views of the cofactors. (A) The electron density associated with the cofactors and the CxxxCxxC motif, conserved in all Radical SAM proteins. The 4Fe–4S cluster is rendered in green (Fe) and yellow (S), while pink-colored bonds highlight SAM1 and SAM2. Both (S)- (above) and (R)- (below) sulfonium sulfur configurations are observed for SAM1. SAM2 is rotationally disordered around the C5′–S+ bond, resulting in discontinuous electron density. (B) The cofactors occupy the central void of the catalytic domain near the C-terminal ends of the three-quarter barrel β-strands. Orange spheres mark the Cα positions of conserved cysteines. (C) A schematic depiction of inter-cofactor distances and amino acid residues involved in binding the cofactors. The (S)-sulfur is presented in yellow and the (R)-sulfur in orange. Green arcs represent hydrophobic interactions.
Fig. 3. Detailed views of the cofactors. (A) The electron density associated with the cofactors and the CxxxCxxC motif, conserved in all Radical SAM proteins. The 4Fe–4S cluster is rendered in green (Fe) and yellow (S), while pink-colored bonds highlight SAM1 and SAM2. Both (S)- (above) and (R)- (below) sulfonium sulfur configurations are observed for SAM1. SAM2 is rotationally disordered around the C5′–S+ bond, resulting in discontinuous electron density. (B) The cofactors occupy the central void of the catalytic domain near the C-terminal ends of the three-quarter barrel β-strands. Orange spheres mark the Cα positions of conserved cysteines. (C) A schematic depiction of inter-cofactor distances and amino acid residues involved in binding the cofactors. The (S)-sulfur is presented in yellow and the (R)-sulfur in orange. Green arcs represent hydrophobic interactions.
Fig. 3. Detailed views of the cofactors. (A) The electron density associated with the cofactors and the CxxxCxxC motif, conserved in all Radical SAM proteins. The 4Fe–4S cluster is rendered in green (Fe) and yellow (S), while pink-colored bonds highlight SAM1 and SAM2. Both (S)- (above) and (R)- (below) sulfonium sulfur configurations are observed for SAM1. SAM2 is rotationally disordered around the C5′–S+ bond, resulting in discontinuous electron density. (B) The cofactors occupy the central void of the catalytic domain near the C-terminal ends of the three-quarter barrel β-strands. Orange spheres mark the Cα positions of conserved cysteines. (C) A schematic depiction of inter-cofactor distances and amino acid residues involved in binding the cofactors. The (S)-sulfur is presented in yellow and the (R)-sulfur in orange. Green arcs represent hydrophobic interactions.
Fig. 4. Distribution of conserved residues in HemN. (A) Amino acid sequence of E.coli HemN. Residues conserved in 34 sequences of HemN are underlaid in dark pink; residues conserved in >90, 80 and 70%, respectively, of sequences are marked by progressively lighter shades. α-Helices are represented by rectangles, β-strands by arrows. See Figure 2 for color-coding and nomenclature. Filled circles, squares and inverted triangles below individual residues mark amino acids involved in 4Fe–4S cluster, SAM1 and SAM2 binding, respectively. Filled squares denote residues in domain–domain interactions. Residues postulated to be involved in binding the external electron donor, terminal electron acceptor and the coproporphyrinogen III substrate are marked by half circles, diamonds and plus signs, respectively, Surface representation of HemN (B) front (in stereo) and (C) back view. The degree of conservation (A) is mapped onto the molecular surface of the catalytic domain. Note that the highest concentration of conserved residues is found in the active site cleft and at domain–domain interfaces. The trip-wire and C-terminal domain are represented by green and red coils. Most of the outer surface is poorly conserved (white), with the exception of the proposed entrance to the terminal electron acceptor-binding pocket and, to a lesser extent, the binding site of the external electron donor.
Fig. 4. Distribution of conserved residues in HemN. (A) Amino acid sequence of E.coli HemN. Residues conserved in 34 sequences of HemN are underlaid in dark pink; residues conserved in >90, 80 and 70%, respectively, of sequences are marked by progressively lighter shades. α-Helices are represented by rectangles, β-strands by arrows. See Figure 2 for color-coding and nomenclature. Filled circles, squares and inverted triangles below individual residues mark amino acids involved in 4Fe–4S cluster, SAM1 and SAM2 binding, respectively. Filled squares denote residues in domain–domain interactions. Residues postulated to be involved in binding the external electron donor, terminal electron acceptor and the coproporphyrinogen III substrate are marked by half circles, diamonds and plus signs, respectively, Surface representation of HemN (B) front (in stereo) and (C) back view. The degree of conservation (A) is mapped onto the molecular surface of the catalytic domain. Note that the highest concentration of conserved residues is found in the active site cleft and at domain–domain interfaces. The trip-wire and C-terminal domain are represented by green and red coils. Most of the outer surface is poorly conserved (white), with the exception of the proposed entrance to the terminal electron acceptor-binding pocket and, to a lesser extent, the binding site of the external electron donor.
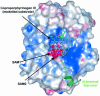
Fig. 5. The modeled substrate (cyan) is depicted in the context of the electrostatic surface potential distribution of the HemN catalytic domain. The binding pockets of SAM1 and SAM2 (pink bonds) are predominantly negatively charged (red), while the proposed substrate-binding pocket is largely positively charged (blue) to accommodate the positively charged sulfonium and the negatively charged propionate side chains, respectively, of coproporphyrinogen III. The C-terminal domain is omitted for clarity, while the N-terminal trip-wire is shown in ribbon-style representation (green). It bears a conserved positively charged residue that may help bind the substrate. This could, in turn, stabilize the trip-wire and tip the C-terminal domain to shut the active site cleft.
Similar articles
- Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines.
Layer G, Grage K, Teschner T, Schünemann V, Breckau D, Masoumi A, Jahn M, Heathcote P, Trautwein AX, Jahn D. Layer G, et al. J Biol Chem. 2005 Aug 12;280(32):29038-46. doi: 10.1074/jbc.M501275200. Epub 2005 Jun 20. J Biol Chem. 2005. PMID: 15967800 - Structure and function of radical SAM enzymes.
Layer G, Heinz DW, Jahn D, Schubert WD. Layer G, et al. Curr Opin Chem Biol. 2004 Oct;8(5):468-76. doi: 10.1016/j.cbpa.2004.08.001. Curr Opin Chem Biol. 2004. PMID: 15450488 Review. - Revisiting the Mechanism of the Anaerobic Coproporphyrinogen III Oxidase HemN.
Ji X, Mo T, Liu WQ, Ding W, Deng Z, Zhang Q. Ji X, et al. Angew Chem Int Ed Engl. 2019 May 6;58(19):6235-6238. doi: 10.1002/anie.201814708. Epub 2019 Apr 1. Angew Chem Int Ed Engl. 2019. PMID: 30884058 - Structural and functional comparison of HemN to other radical SAM enzymes.
Layer G, Kervio E, Morlock G, Heinz DW, Jahn D, Retey J, Schubert WD. Layer G, et al. Biol Chem. 2005 Oct;386(10):971-80. doi: 10.1515/BC.2005.113. Biol Chem. 2005. PMID: 16218869 Review. - Oxygen-independent coproporphyrinogen-III oxidase HemN from Escherichia coli.
Layer G, Verfürth K, Mahlitz E, Jahn D. Layer G, et al. J Biol Chem. 2002 Sep 13;277(37):34136-42. doi: 10.1074/jbc.M205247200. Epub 2002 Jul 11. J Biol Chem. 2002. PMID: 12114526
Cited by
- Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin.
Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips JD. Dailey HA, et al. Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2210-5. doi: 10.1073/pnas.1416285112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646457 Free PMC article. - Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Zhang Y, et al. Nature. 2010 Jun 17;465(7300):891-6. doi: 10.1038/nature09138. Nature. 2010. PMID: 20559380 Free PMC article. - AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation.
Nicolet Y, Drennan CL. Nicolet Y, et al. Nucleic Acids Res. 2004 Aug 2;32(13):4015-25. doi: 10.1093/nar/gkh728. Print 2004. Nucleic Acids Res. 2004. PMID: 15289575 Free PMC article. - Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical _S_-Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
Zhang J, Hou X, Chen Z, Ko Y, Ruszczycky MW, Chen Y, Zhou J, Liu HW. Zhang J, et al. J Am Chem Soc. 2022 Jun 8;144(22):9910-9919. doi: 10.1021/jacs.2c02676. Epub 2022 May 27. J Am Chem Soc. 2022. PMID: 35622017 Free PMC article. - Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme.
Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL. Vey JL, et al. Proc Natl Acad Sci U S A. 2008 Oct 21;105(42):16137-41. doi: 10.1073/pnas.0806640105. Epub 2008 Oct 13. Proc Natl Acad Sci U S A. 2008. PMID: 18852451 Free PMC article.
References
- Becker A., Fritz-Wolf,K., Kabsch,W., Knappe,J., Schultz,S. and Wagner,A.F.V. (1999) Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat. Struct. Biol., 6, 969–975. - PubMed
- Cannon L.M., Butler,F.N., Wan,W. and Zhou,Z.S. (2002) A stereo specific colorimetric assay for (S,S)-adenosylmethionine quantification based on thiopurine methyltransferase-catalyzed, thiol methylation. Anal. Biochem., 308, 358–363. - PubMed
- CCP4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. - PubMed
- Chadwick D.J. and Ackrill,K. (eds) (1994) Ciba Foundation Symposia 180: The Biosynthesis of the Tetrapyrrole Pigments. Wiley and Sons, Chichester, UK, pp. 131–155.
- Cheek J. and Broderick,J.B. (2001) Adenosylmethionine-dependent iron–sulfur enzymes: versatile clusters in a radical new role. J. Biol. Inorg. Chem., 6, 209–226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases