Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene - PubMed (original) (raw)
. 2003 Dec;112(12):1809-20.
doi: 10.1172/JCI20039. Epub 2003 Nov 24.
Jie Yu, Govind Bhagat, Norihiko Furuya, Hanina Hibshoosh, Andrea Troxel, Jeffrey Rosen, Eeva-Liisa Eskelinen, Noboru Mizushima, Yoshinori Ohsumi, Giorgio Cattoretti, Beth Levine
Affiliations
- PMID: 14638851
- PMCID: PMC297002
- DOI: 10.1172/JCI20039
Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene
Xueping Qu et al. J Clin Invest. 2003 Dec.
Abstract
Malignant cells often display defects in autophagy, an evolutionarily conserved pathway for degrading long-lived proteins and cytoplasmic organelles. However, as yet, there is no genetic evidence for a role of autophagy genes in tumor suppression. The beclin 1 autophagy gene is monoallelically deleted in 40-75% of cases of human sporadic breast, ovarian, and prostate cancer. Therefore, we used a targeted mutant mouse model to test the hypothesis that monoallelic deletion of beclin 1 promotes tumorigenesis. Here we show that heterozygous disruption of beclin 1 increases the frequency of spontaneous malignancies and accelerates the development of hepatitis B virus-induced premalignant lesions. Molecular analyses of tumors in beclin 1 heterozygous mice show that the remaining wild-type allele is neither mutated nor silenced. Furthermore, beclin 1 heterozygous disruption results in increased cellular proliferation and reduced autophagy in vivo. These findings demonstrate that beclin 1 is a haplo-insufficient tumor-suppressor gene and provide genetic evidence that autophagy is a novel mechanism of cell-growth control and tumor suppression. Thus, mutation of beclin 1 or other autophagy genes may contribute to the pathogenesis of human cancers.
Figures
Figure 1
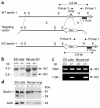
Targeted disruption of beclin 1 in mice. (a) Restriction maps of the wild-type beclin 1 allele (top), the beclin 1 targeting vector (middle), and the predicted targeted beclin 1 allele (bottom). Restriction sites are as follows: EcoRI (E), HindIII (H). The targeting construct contains a cassette with the neomycin resistance gene (Neo) that has replaced exons 1 and 2 of the beclin 1 gene. “X” denotes regions of homologous recombination between the targeting vector and wild-type allele. The beclin 1 genomic fragment used as a 3′ external probe for Southern blot analysis is indicated by a solid black box. Expected sizes of the EcoRI fragments that hybridize with the probe are indicated. (b) Southern blot analysis of genomic DNA from beclin 1+/+ and beclin 1+/– ES cells and mouse tails. The DNA was digested with EcoRI and hybridized with the probe indicated in a. The sizes of wild-type (WT) and disrupted (KO) alleles are shown. (c) PCR genotyping of genomic DNA from beclin 1+/– and beclin 1+/+ ES cells and mouse tail DNA. Primers 1 and 2 in a were used to detect the wild-type allele, and primers 1 and 3 in a were used to detect the knockout allele. (d) Western blot analysis of Beclin 1 protein expression in beclin 1+/+ and beclin 1+/– ES cells and mouse lung samples. Sizes of Beclin 1 isoforms and an actin control are indicated on the right. Lung lysates were prepared from 2-month-old mice. Similar results were observed for samples from six different mice of each genotype.
Figure 2
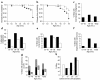
Increase in the frequency of spontaneous malignancies (a–g) and accelerated development of HBV-induced premalignant lesions in beclin 1 heterozygous-deficient mice (h). (a and b) Kaplan-Meier plot of time to development of macroscopic malignancy (a) and any malignancy (b) in beclin 1+/– (solid lines, filled circles) versus beclin 1+/+ (dotted lines, open circles) mice (P < 0.0001, log-rank test). “Macroscopic malignancy” refers to tumors observed upon gross inspection that were subsequently confirmed to be malignancies upon histologic examination. “Any malignancy” denotes either a macroscopic or a microscopic malignancy detected upon complete histologic survey of all major internal organs. (**c**–**g**) Prevalence of macroscopic malignancies (**c**), all malignancies (**d**), lung carcinomas (**e**), hepatocellular carcinomas (**f**), and lymphomas and lymphoproliferative disease (LPD; **g**) in _beclin 1+/–_ (black bars) versus _beclin 1+/+_ (white bars) mice (_P_ < 0.0001, Fisher’s exact test for **c**–**g**). In **g**, black denotes lymphoma and gray denotes lymphoproliferative disease. (**h**) Extent of small-cell dysplasia in livers from 13-month-old _beclin 1+/–_ (_n_ = 27; black bars) versus _beclin 1+/+_ (_n_ = 32; white bars) HBV transgenic mice. The scale for small-cell dysplasia (72) is: 0, absent or rare foci; 1+, <25% of liver with small-cell dysplasia; 2+, 25–50% of liver with small-cell dysplasia; 3+, >50% of liver with small-cell dysplasia. Beclin 1+/– HBV transgenic mice have significantly more severe disease (P = 0.0289, Mantel-Haenszel χ2 test).
Figure 3
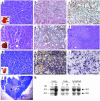
Histopathologic and molecular characterization of lesions in beclin 1+/– mice. (a–c) Representative well-differentiated papillary lung carcinoma in a beclin 1+/– mouse, stained with H&E (a), anti–Beclin 1 (b), or anti–TTF-1 (c). The inset in a shows lungs with subpleural tumor nodules (arrows). Beclin 1 immunoreactivity levels similar to those shown in b were observed in all lung carcinomas in beclin 1+/– mice (n = 10; data not shown). (d and e) Representative well-differentiated hepatocellular carcinoma in a beclin 1+/– mouse, stained with H&E (d) or anti–Beclin 1 (e). The inset in d shows gross pathology of liver tumor. Beclin 1 immunoreactivity levels similar to those shown in e were observed in all hepatocellular carcinomas identified in beclin 1+/– mice (n = 4; data not shown). (f) Representative area of preneoplastic small-cell dysplasia in the liver of a beclin 1+/– mouse that transgenically expresses the HBV large-envelope polypeptide. (g–i) Representative DLCL in a beclin 1+/– mouse, stained with H&E (g), anti-Pax5 (dark purple) and anti-CD3 (brown; h), and anti–BCL-6 (i). The inset in g shows lymphoma (arrow) adjacent to normal kidney. (j) Example of lymphoproliferative disease in the thymus of a beclin 1+/– mouse with medullary follicular hyperplasia and cortical effacement. The arrow denotes a germinal center. (k) Southern blot analysis to detect the wild-type and disrupted beclin 1 allele in tumor samples and matched normal tissue. No evidence of allelic loss is seen in beclin 1+/– tumors. Data are shown for one example of each of the three types of malignancy observed. Similar findings were observed for all palpable malignancies (n = 15). Scale bars: a–i, 50 μm; j, 1 mm.
Figure 4
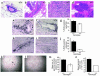
Beclin 1 heterozygous deletion alters cell-growth control in the mammary gland and in splenic germinal centers. (a–d) H&E sections of normal mammary epithelial duct in a beclin 1+/+ mouse (a) and hyperplastic/neoplastic lesions in the mammary glands of beclin 1+/– 6- to 9-month-old virgin mice (b–d), including mammary intraepithelial neoplasia (b), adenomyoepithelioma (c), and acinar neoplasia (d). (e–j) Representative images of BrdU staining of terminal end buds (TEBs) (e and f) and mammary ducts (h and i) in 5-week-old virgin mice and quantitation of percentage of BrdU-positive cells in TEBs (g) and mammary ducts (j). Results shown represent the mean ± SEM of 15 TEBs (g) or 15 ducts (j) from five mice (aged 5 weeks) per genotype. Significant differences were observed between beclin 1+/– and beclin 1+/+ genotypes for percentages of both BrdU-positive TEB cells (P = 0.025, t test) and BrdU-positive ductal cells (P < 0.0001, t test). The arrow in e indicates the neck region, and the arrow in f indicates the cap cell region. (k–n) Representative images of splenic germinal centers (arrow) labeled by PNA staining (38) 8 days after immunization with sheep rbc’s (k and l), and quantitation of the number (m) and size (n) of germinal centers. Results shown represent the mean ± SEM from spleen sections analyzed from five mice per genotype. Significant differences were observed between beclin 1+/– and beclin 1+/+ genotypes for both number (P = 0.027, t test) and size (P = 0.018, t test) of germinal centers. Scale bars: a–f, h, and i, 100 μm; k and l, 1 mm.
Figure 5
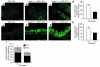
Beclin 1 heterozygous deletion decreases autophagy in muscle (a–d), bronchial epithelia (e–h), and germinal center B lymphocytes (i). (a–d) Representative images of GFP-LC3 staining in (a–c) and quantitation of GFP-LC3–positive dots (d) in muscle from 2-month-old mice subjected to 24-hour starvation. The x axis labels denote beclin 1 genotype. Results shown in d represent the mean ± SEM for approximately 50 images obtained from five mice per genotype (P < 0.0001, t test). (e–h) Representative images of GFP-LC3 staining (e–g) and quantitation of GFP-LC3–positive dots (h) in bronchial epithelial tissue from 2-month-old mice subjected to 24-hour starvation. The x axis labels denote beclin 1 genotype. Results shown in h represent the mean ± SEM for approximately 50 images obtained from five mice per genotype (P < 0.0001, t test). (i) Morphometric electron microscopic quantitation of autophagic vacuole (AV) profiles in germinal center B lymphocytes isolated from 2-month-old nonstarved mice. The x axis labels denote beclin 1 genotype. Avi denotes early, immature autophagic vacuoles, and Avd denotes late, degradative autophagic vacuoles. Results shown in i represent the mean ± SEM for parallel quantitations from seven to eight grid squares (P = 0.02, t test). Arrows in b and f denote representative GFP-LC3–positive punctate dots that are quantitated in d and h, respectively. Scale bars: 10 nm.
Comment in
- The importance of 'self-eating'.
Wrighton KH. Wrighton KH. Nat Rev Mol Cell Biol. 2010 Oct;11(10):681. doi: 10.1038/nrm2978. Nat Rev Mol Cell Biol. 2010. PMID: 20861878 No abstract available.
Similar articles
- Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor.
Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Yue Z, et al. Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):15077-82. doi: 10.1073/pnas.2436255100. Epub 2003 Dec 1. Proc Natl Acad Sci U S A. 2003. PMID: 14657337 Free PMC article. - Induction of autophagy and inhibition of tumorigenesis by beclin 1.
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Liang XH, et al. Nature. 1999 Dec 9;402(6762):672-6. doi: 10.1038/45257. Nature. 1999. PMID: 10604474 - Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function.
Liang XH, Yu J, Brown K, Levine B. Liang XH, et al. Cancer Res. 2001 Apr 15;61(8):3443-9. Cancer Res. 2001. PMID: 11309306 - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review. - Role and regulation of autophagy in cancer.
Chen N, Karantza-Wadsworth V. Chen N, et al. Biochim Biophys Acta. 2009 Sep;1793(9):1516-23. doi: 10.1016/j.bbamcr.2008.12.013. Epub 2009 Jan 2. Biochim Biophys Acta. 2009. PMID: 19167434 Free PMC article. Review.
Cited by
- Anti-Cancer Strategy Based on Changes in the Role of Autophagy Depending on the Survival Environment and Tumorigenesis Stages.
Lee M, Kim HG. Lee M, et al. Molecules. 2024 Oct 30;29(21):5134. doi: 10.3390/molecules29215134. Molecules. 2024. PMID: 39519774 Free PMC article. Review. - Selective protein degradation through chaperone‑mediated autophagy: Implications for cellular homeostasis and disease (Review).
Huang J, Wang J. Huang J, et al. Mol Med Rep. 2025 Jan;31(1):13. doi: 10.3892/mmr.2024.13378. Epub 2024 Nov 8. Mol Med Rep. 2025. PMID: 39513615 Free PMC article. Review. - Mitoepigenetics pathways and natural compounds: a dual approach to combatting hepatocellular carcinoma.
Hatawsh A, Al-Haddad RH, Okafor UG, Diab LM, Dekanoidze N, Abdulwahab AA, Mohammed OA, Doghish AS, Moussa R, Elimam H. Hatawsh A, et al. Med Oncol. 2024 Oct 27;41(12):302. doi: 10.1007/s12032-024-02538-8. Med Oncol. 2024. PMID: 39465473 Review. - A Four-Gene Autophagy-Related Prognostic Model Signature and Its Association With Immune Phenotype in Lung Squamous Cell Carcinoma.
Luo L, Deng J, Tang Q. Luo L, et al. Cancer Rep (Hoboken). 2024 Oct;7(10):e70000. doi: 10.1002/cnr2.70000. Cancer Rep (Hoboken). 2024. PMID: 39443755 Free PMC article. - Promising and challenging phytochemicals targeting LC3 mediated autophagy signaling in cancer therapy.
Rajendran P, Renu K, Ali EM, Genena MAM, Veeraraghavan V, Sekar R, Sekar AK, Tejavat S, Barik P, Abdallah BM. Rajendran P, et al. Immun Inflamm Dis. 2024 Oct;12(10):e70041. doi: 10.1002/iid3.70041. Immun Inflamm Dis. 2024. PMID: 39436197 Free PMC article. Review.
References
- Elliott PJ, Ross JS. The proteasome: a new target for novel drug therapies. Am. J. Clin. Pathol. 2001;116:637–646. - PubMed
- Richardson P. Clinical update: proteasome inhibitors in hematologic malignancies. Cancer Treat. Rev. 2003;29(Suppl. 1):33–39. - PubMed
- Lenz HJ. Clinical update: proteasome inhibitors in solid tumors. Cancer Treat. Rev. 2003;29(Suppl. 1):41–48. - PubMed
- Bursch W, et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MC7-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA084254/CA/NCI NIH HHS/United States
- R01 CA16303/CA/NCI NIH HHS/United States
- R01 CA84254/CA/NCI NIH HHS/United States
- R01 AI44157/AI/NIAID NIH HHS/United States
- R01 CA016303/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials