Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3 - PubMed (original) (raw)
Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3
Andries Blokzijl et al. J Cell Biol. 2003.
Abstract
The Notch and transforming growth factor-beta (TGF-beta) signaling pathways play critical roles in the control of cell fate during metazoan development. However, mechanisms of cross-talk and signal integration between the two systems are unknown. Here, we demonstrate a functional synergism between Notch and TGF-beta signaling in the regulation of Hes-1, a direct target of the Notch pathway. Activation of TGF-beta signaling up-regulated Hes-1 expression in vitro and in vivo. This effect was abrogated in myogenic cells by a dominant-negative form of CSL, an essential DNA-binding component of the Notch pathway. TGF-beta regulated transcription from the Hes-1 promoter in a Notch-dependent manner, and the intracellular domain of Notch1 (NICD) cooperated synergistically with Smad3, an intracellular transducer of TGF-beta signals, to induce the activation of synthetic promoters containing multimerized CSL- or Smad3-binding sites. NICD and Smad3 were shown to interact directly, both in vitro and in cells, in a ligand-dependent manner, and Smad3 could be recruited to CSL-binding sites on DNA in the presence of CSL and NICD. These findings indicate that Notch and TGF-beta signals are integrated by direct protein-protein interactions between the signal-transducing intracellular elements from both pathways.
Figures
Figure 1.
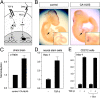
**Induction of Hes-1 expression by TGF-**β signaling in vivo and in cell culture. (A) Scheme summarizing principal similarities in membrane-to-nucleus signaling between the Notch and TGF-β signaling pathways. (B) Induction of c-hairy mRNA expression in embryonic chick heart (arrows) by TGF-β signaling. Images show whole-mount in situ hybridization c-hairy mRNA of E4 chick embryos electroporated with GFP (control) or GFP plus CA-ALK5 expression plasmids. Induction of c-hairy mRNA expression could be observed in six out of six embryos electroporated with CA-ALK5. In no case (four out of four embryos) was c-hairy mRNA expression observed in control embryos. (C) Real-time PCR analysis of c-hairy mRNA expression in electroporated embryonic chick brain. The histogram shows means of three independent experiments each performed in triplicate ± SEM. (D) Real-time PCR analysis of Hes-1 mRNA expression in adult mouse neural stem cells treated with 10 ng/ml TGF-β for 90 min before RNA extraction. Results are presented as the mean ± SD. (E) Real-time PCR analysis of Hes-1 mRNA expression in C2C12 mouse myoblasts treated with 10 ng/ml TGF-β for 60 min before RNA extraction. Cycloheximide treatment (+ Chx) was begun 10 min before TGF-β stimulation. Results are presented as the mean ± SD of triplicate determinations.
Figure 2.
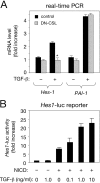
**Requirement of Notch signaling for the effects of TGF-**β on Hes-1 expression. (A) Real-time PCR analysis of Hes-1 mRNA expression in C2C12 mouse myoblasts expressing a dominant-negative CSL construct (DN-CSL) after treatment with TGF-β. Cells expressing DN-CSL (cotransfected with a GFP construct) were selected by FACS® analysis and treated with TGF-β for 60 min before RNA extraction. Results are presented as the mean ± SD of triplicate determinations. (B) Activation of the Hes-1 promoter in C2C12 myoblasts by NICD and TGF-β. A 500-bp construct of the Hes-1 promoter coupled to a luciferase reporter was introduced into C2C12 cells along with NICD (from mouse Notch1) as indicated. Treatment with TGF-β resulted in potentiation of NICD activity in a dose-dependent manner. Results are presented as the mean ± SD of triplicate determinations.
Figure 3.
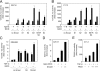
Cooperation of NICD and Smad3 in the activation of Notch- and Smad-specific synthetic promoters. (A) Role of individual Smad proteins in the regulation of the 12xCSL-luc reporter by NICD and TGF-β ligands in C2C12 myoblasts. Smad expression plasmids as indicated were transfected along with the 12xCSL-luc reporter construct in the presence or absence of NICD and TGF-β ligands at 10 ng/ml. T, TGF-β1; B, BMP-4. Normalized results are expressed relative to control as the mean ± SD of triplicate determinations. (B) Role of individual Smad proteins in the regulation of the 12xCSL-luc reporter by NICD and TGF-β ligands in C17.2 neural stem cells. (C) Activation of the 12xCSL-luc reporter in MDA468 breast cancer cells (lacking Smad4) by NICD and Smad3. (D) Activation of the Gal4-luc reporter in C2C12 myoblasts by Gal4–NICD and Smad3. (E) Activation of the Smad3-specific CAGA-luc reporter in C17.2 neural stem cells by TGF-β and potentiation by increasing amounts of NICD.
Figure 4.
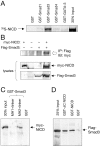
Physical interaction between NICD and Smad3. (A) Full-length 35S-labeled NICD produced by in vitro translation was used in precipitation assays together with equal amounts of the indicated GST–Smad fusion proteins. A GST fusion of the zinc-finger transcription factor GATA-3 was used as a negative control. (B) Coimmunoprecipitation of myc-NICD and Flag-Smad3 in total lysates of transfected COS cells. The anti-Flag antibody directed against Flag-tagged Smad3 brings down myc-tagged NICD only in cells that also received the Flag-tagged Smad3 construct (top). The panels below show immunoblots of 20% of the lysates. (C) In vitro pull-down of myc-tagged NICD from COS cell lysates with GST fusions of the MH1 and MH2 domains of Smad3, including the intervening linker region. (D) In vitro pull-down of Flag-tagged Smad3 from COS cell lysates using GST fusions of full-length NICD or a COOH-terminally truncated NICD construct lacking the transactivation and p300/CBP-binding domains (ΔC NICD).
Figure 5.
Ligand-dependent interaction between endogenous Smad3 and Notch, and recruitment of Smad3 to specific DNA sites by CSL and NICD. (A) Parental C2C12 cells or a stable C2C12 transfectant overexpressing full-length Notch1 (C2C12-N1) were either left untreated or stimulated with 10 ng/ml TGF-β1 for 50 min before lysis and immunoprecipitation with anti-Notch1 antibodies. Immunoblots were probed with anti-Smad3 antibodies and reprobed with anti-Notch1, detected as its transmembrane and intracellular (TMIC) domain. It should be noted that to date, nuclear NICD has been very difficult to detect by biochemical or in situ methods in normal cells, possibly because it is present in very low amounts and/or has a very short half-life (Rand et al., 2000). (B) A biotinylated oligonucleotide containing two tandem CSL-binding sites (2xCSL) was used to pull down GST–Smad3, 35S-labeled NICD, and 35S-labeled CSL in different combinations as indicated. A mutant oligonucleotide (mut) was used as a control. Note that GST–Smad3 (detected by immunoblot with anti-GST antibodies) could only be recovered using the wild-type (wt) oligonucleotide in the presence of both NICD and CSL. Weak levels of 35S-labeled NICD could also be detected in the same lane. (C) A mechanism for the integration of TGF-β and Notch signaling by direct interaction between Smad3 and NICD. Although the Smad3–Smad4 complex and the NICD can translocate to the nucleus independently, our results do not rule out the possibility that their interaction could already take place in the cytoplasm.
Similar articles
- Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling.
Sun Y, Lowther W, Kato K, Bianco C, Kenney N, Strizzi L, Raafat D, Hirota M, Khan NI, Bargo S, Jones B, Salomon D, Callahan R. Sun Y, et al. Oncogene. 2005 Aug 11;24(34):5365-74. doi: 10.1038/sj.onc.1208528. Oncogene. 2005. PMID: 16007227 - Smad3/AP-1 interactions control transcriptional responses to TGF-beta in a promoter-specific manner.
Verrecchia F, Vindevoghel L, Lechleider RJ, Uitto J, Roberts AB, Mauviel A. Verrecchia F, et al. Oncogene. 2001 Jun 7;20(26):3332-40. doi: 10.1038/sj.onc.1204448. Oncogene. 2001. PMID: 11423983 - Retinoic acid receptors interfere with the TGF-beta/Smad signaling pathway in a ligand-specific manner.
Pendaries V, Verrecchia F, Michel S, Mauviel A. Pendaries V, et al. Oncogene. 2003 Nov 6;22(50):8212-20. doi: 10.1038/sj.onc.1206913. Oncogene. 2003. PMID: 14603262 - HES and HERP families: multiple effectors of the Notch signaling pathway.
Iso T, Kedes L, Hamamori Y. Iso T, et al. J Cell Physiol. 2003 Mar;194(3):237-55. doi: 10.1002/jcp.10208. J Cell Physiol. 2003. PMID: 12548545 Review. - The role of transcription factors involved in TGFbeta superfamily signaling during development.
Watanabe M, Whitman M. Watanabe M, et al. Cell Mol Biol (Noisy-le-grand). 1999 Jul;45(5):537-43. Cell Mol Biol (Noisy-le-grand). 1999. PMID: 10512186 Review.
Cited by
- Loss of TGF-β adaptor β2SP activates notch signaling and SOX9 expression in esophageal adenocarcinoma.
Song S, Maru DM, Ajani JA, Chan CH, Honjo S, Lin HK, Correa A, Hofstetter WL, Davila M, Stroehlein J, Mishra L. Song S, et al. Cancer Res. 2013 Apr 1;73(7):2159-69. doi: 10.1158/0008-5472.CAN-12-1962. Epub 2013 Mar 27. Cancer Res. 2013. PMID: 23536563 Free PMC article. - Molecular pathways of notch signaling in vascular smooth muscle cells.
Boucher J, Gridley T, Liaw L. Boucher J, et al. Front Physiol. 2012 Apr 9;3:81. doi: 10.3389/fphys.2012.00081. eCollection 2012. Front Physiol. 2012. PMID: 22509166 Free PMC article. - Notch3 signaling promotes the development of pulmonary arterial hypertension.
Li X, Zhang X, Leathers R, Makino A, Huang C, Parsa P, Macias J, Yuan JX, Jamieson SW, Thistlethwaite PA. Li X, et al. Nat Med. 2009 Nov;15(11):1289-97. doi: 10.1038/nm.2021. Epub 2009 Oct 25. Nat Med. 2009. PMID: 19855400 Free PMC article. - RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development.
Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, O'Keefe RJ, Hilton MJ. Dong Y, et al. Development. 2010 May;137(9):1461-71. doi: 10.1242/dev.042911. Epub 2010 Mar 24. Development. 2010. PMID: 20335360 Free PMC article. - Metalloprotease-disintegrin ADAM12 expression is regulated by Notch signaling via microRNA-29.
Li H, Solomon E, Duhachek Muggy S, Sun D, Zolkiewska A. Li H, et al. J Biol Chem. 2011 Jun 17;286(24):21500-10. doi: 10.1074/jbc.M110.207951. Epub 2011 Apr 25. J Biol Chem. 2011. PMID: 21518768 Free PMC article.
References
- Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. - PubMed
- Attisano, L., and J.L. Wrana. 2002. Signal transduction by the TGF-β superfamily. Science. 296:1646–1647. - PubMed
- Beatus, P., J. Lundkvist, C. Oberg, K. Pedersen, and U. Lendahl. 2001. The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity. Mech. Dev. 104:3–20. - PubMed
- Blokzijl, A., P. ten Dijke, and C.F. Ibáñez. 2002. Physical and functional interaction between GATA-3 and Smad3 allows TGF-β regulation of GATA target genes. Curr. Biol. 12:35–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources