Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2 - PubMed (original) (raw)
Comparative Study
. 2003 Dec;73(6):1302-15.
doi: 10.1086/379979. Epub 2003 Nov 24.
Affiliations
- PMID: 14639526
- PMCID: PMC1180396
- DOI: 10.1086/379979
Comparative Study
Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2
Weimin Bi et al. Am J Hum Genet. 2003 Dec.
Abstract
Smith-Magenis syndrome (SMS) is caused by an approximately 4-Mb heterozygous interstitial deletion on chromosome 17p11.2 in approximately 80%-90% of affected patients. Three large ( approximately 200 kb), complex, and highly homologous ( approximately 98%) low-copy repeats (LCRs) are located inside or flanking the SMS common deletion. These repeats, also known as "SMS-REPs," are termed "distal," "middle," and "proximal." The directly oriented distal and proximal copies act as substrates for nonallelic homologous recombination resulting in both the deletion associated with SMS and the reciprocal duplication: dup(17)(p11.2p11.2). Using restriction enzyme cis-morphism analyses and direct sequencing, we mapped the regions of strand exchange in 16 somatic-cell hybrids that harbor only the recombinant SMS-REP. Our studies showed that the sites of crossovers were distributed throughout the region of homology between the distal and proximal SMS-REPs. However, despite approximately 170 kb of high homology, 50% of the recombinant junctions occurred in a 12.0-kb region within the KER gene clusters. DNA sequencing of this hotspot (positional preference for strand exchange) in seven recombinant SMS-REPs narrowed the crossovers to an approximately 8-kb interval. Four of them occurred in a 1,655-bp region rich in polymorphic nucleotides that could potentially reflect frequent gene conversion. For further evaluation of the strand exchange frequency in patients with SMS, novel junction fragments from the recombinant SMS-REPs were identified. As predicted by the reciprocal-recombination model, junction fragments were also identified from this hotspot region in patients with dup(17)(p11.2p11.2), documenting reciprocity of the positional preference for strand exchange. Several potential cis-acting recombination-promoting sequences were identified within the hotspot. It is interesting that we found 2.1-kb AT-rich inverted repeats flanking the proximal and middle KER gene clusters but not the distal one. The role of any or all of these in stimulating double-strand breaks around this positional recombination hotspot remains to be explored.
Figures
Figure 1
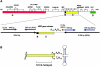
Refining the regions of unequal crossover in somatic-cell hybrids. Top, the genomic structures of SMS-REPs, with the distal and proximal copies in direct orientation, telomere (tel) (left) and centromere (cen) (right). Shown above each SMS-REP copy are the pseudogenes (unblackened horizontal rectangles) and the direction of transcription (arrows). Shown below the SMS-REPs are the homology segments. As described elsewhere (Park et al. 2002), the A, B, C, and D sequence blocks have >98% identity between the distal (unblackened rectangles) and the proximal (hatched rectangles) SMS-REP. Two segments (horizontal brackets) in the proximal SMS-REP (∼47 kb between blocks A and B, and ∼39 kb between blocks C and D) are absent in the distal copy. Bottom, regions of strand exchange within the recombinant SMS-REP of the deletion chromosome isolated in hybrids. Restriction _cis_-morphism markers enabling distinction between the distal and proximal SMS-REP copies are indicated (circles), with their positions corresponding to the proximal SMS-REP at the top of the figure. Some sites in the recombinant SMS-REP were derived from the distal SMS-REP (unblackened circles), and others were derived from the proximal copy (hatched circles). For each somatic-cell hybrid, the region of strand exchange within the recombinant SMS-REP and its size are indicated (blackened horizontal bar). The recombination event in hybrid 255-11D is centromeric to the CLP region (D) in the distal SMS-REP.
Figure 2
Sites of strand exchanges in recombinant SMS-REPs in somatic-cell hybrids. Top, the nucleotide differences between the proximal and distal SMS-REPs. The positions of the first and the last nucleotide difference in the 8.7-kb recombinant region of both proximal and distal SMS-REP copies are shown (see text for details). The absence of the corresponding nucleotide is denoted by a vertical dotted line. A total of 39 differences in DNA sequence (uppercase letters) between proximal and distal copies of SMS-REP were used for mapping the sites of strand exchange. These represent PSVs, also known as “_cis_-morphisms” (Lupski 2003), between proximal and distal copies and thus enable distinctions between the copies. Twenty-five sites are polymorphic (lowercase letters) and thus noninformative (small shaded circle) for mapping recombinants. The recombinant SMS-REP of each individual hybrid (horizontal line) is shown, with the hybrid number given at left. The DNA sequence differences between the distal and proximal SMS-REP are represented by circles for ease of analysis; white circles denote matches to the distal SMS-REP, and hatched circles denote matches to the proximal SMS-REP. The _cis_-morphic restriction site (_Bam_HI) used for the identification of novel junction fragments and its genomic position (in parentheses) in the distal SMS-REP are indicated. regions of strand exchange in each hybrid are highlighted using a thick black line. The dotted square on the recombinant chromosome from 283-15D reveals an apparent gene-conversion event. Vertical dotted lines demarcate regions of increased polymorphic variation, potentially occurring because of gene conversion accompany crossovers, that overlap with a positional preference for unequal crossovers. Bottom, summary of regions of strand exchange in each hybrid (blackened rectangles), with the nucleotide length given above. The genomic sequence positions of these strand exchange regions are marked below according to the nucleotide numbers in the distal SMS-REP.
Figure 3
Junction fragments identified in patients with SMS and dup(17)(p11.2p11.2). A, sequence prediction of novel junction fragments from recombinant SMS-REPs. Portions of the KER gene cluster of distal (dist) and proximal (prox) SMS-REPs are shown with the horizontal dotted and hatched rectangles depicting homology segments. A 9.7-kb fragment from the distal SMS-REP and an 11.5-kb fragment from the proximal SMS-REP are predicted when genomic DNA is double-digested with _Bam_HI and _Spe_I and hybridized with a 906-bp probe (short horizontal bar). In addition, a 6.9-kb junction fragment from patients with the SMS deletion and a 14.5-kb junction fragment in its reciprocal duplication from patients with dup(17)(p11.2p11.2) are predicted. B, DNA samples from patients (468 and 244) and del(17)p11.2 retaining hybrids were digested with _Bam_HI and _Spe_I. A predicted 6.9-kb novel junction fragment band was observed in patient 468 and in hybrids 363-1D and 468-5D. Hybrid 255-11D had only the band from the distal SMS-REP, whereas hybrids 254-1D, 280-22D, 251-20D, and 244-2D had the band from the proximal SMS-REP. C, The 6.9-kb novel junction fragment cosegregates with the SMS phenotype. Only the affected individuals (blackened symbols) and neither parent (in the case of families) had the novel 6.9-kb junction fragment. The three control lanes contain BAC DNA and indicate that the 9.7-kb band was from the distal (RP11-219A15) SMS-REP, whereas both the middle (RP11-158M20) and proximal (RP11-434D2) SMS-REPs generated an 11.5-kb band. D, Identification of a novel junction fragment in patients with dup(17)(p11.2p11.2). DNA samples from three families were digested with _Bam_HI and _Spe_1. A 14.5-kb novel junction fragment was observed only in patients, not in their unaffected family members. Note an ∼16-kb band is nonspecific to SMS-REPs.
Figure 4
Distribution of sequence blocks with perfect identity between the distal and proximal SMS-REPs. Regions with 200–300 bp perfect identity (shaded blocks) and regions with >300 bp perfect identity (blackened blocks) are shown with the size of identity in base pairs. The four homology blocks between the distal and proximal SMS-REPs are denoted as blocks A (red), B (black), C (yellow), and D (green). The two unequal crossover preferential sites are delineated by horizontal double-end arrows, with the occurrence rate and size indicated. One of them is a 12-kb region at the centromeric end of the homology block C, where 50% (8/16) of strand exchanges occurred. By use of both hybrid analyses and Southern blotting, 15% (5/34) exchanges were mapped in an ∼6.9-kb interval. The 1,655-bp region within the 12-kb hotspot is responsible for four strand exchanges among 16 hybrids. Another hotspot is an ∼1.1-kb region at the centromeric end of the homology block A that accounts for 19% (3/16) of the unequal crossovers. The locations of χ sites (blue arrows), human minisatellite core sequences (black arrows), and a human hypervariable minisatellite (purple arrow) are indicated.
Figure 5
Repeats flanking the KER gene cluster. A, The 2.1-kb AT-rich (38% GC) inverted repeats (black arrows) with 98% homology flank the sequence blocks B and C, including the KER gene cluster, in both the proximal and the middle SMS-REPs, but not in the distal SMS-REP. Note that the sizes of the homology blocks and the flanking inverted repeats are not drawn to scale. Within the inverted repeat, there are 646-bp internal inverted repeats with 93% homology (blue arrows). One repeat centromeric to the block C is separated from block C with a 91-bp segment (brown arrows) that shares 98% homology with a segment within the inverted repeat and 17 additional nucleotides, including 4 adenines (indicated as A4N13). The 4 adenines are within a tract of 19 continuous adenines, in which 15 are located within block C, the homologous region between the distal and proximal SMS-REPs. B, Each of the inverted repeats can theoretically form hairpin structures as indicated. The 12-kb hotspot region is adjacent to the inverted repeat centromeric to the block C. The long adenine tract and the hairpin structures may be more sensitive to DSBs, which might serve as the initiation event for NAHR.
Similar articles
- Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates.
Shaw CJ, Withers MA, Lupski JR. Shaw CJ, et al. Am J Hum Genet. 2004 Jul;75(1):75-81. doi: 10.1086/422016. Epub 2004 May 13. Am J Hum Genet. 2004. PMID: 15148657 Free PMC article. - Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2.
Shaw CJ, Bi W, Lupski JR. Shaw CJ, et al. Am J Hum Genet. 2002 Nov;71(5):1072-81. doi: 10.1086/344346. Epub 2002 Oct 9. Am J Hum Genet. 2002. PMID: 12375235 Free PMC article. - Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs.
Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR. Park SS, et al. Genome Res. 2002 May;12(5):729-38. doi: 10.1101/gr.82802. Genome Res. 2002. PMID: 11997339 Free PMC article. - Trisomy 17p10-p12 due to mosaic supernumerary marker chromosome: delineation of molecular breakpoints and clinical phenotype, and comparison to other proximal 17p segmental duplications.
Yatsenko SA, Treadwell-Deering D, Krull K, Lewis RA, Glaze D, Stankiewicz P, Lupski JR, Potocki L. Yatsenko SA, et al. Am J Med Genet A. 2005 Oct 1;138A(2):175-80. doi: 10.1002/ajmg.a.30948. Am J Med Genet A. 2005. PMID: 16152635 Review. - Behavioral phenotype of Smith-Magenis syndrome (del 17p11.2).
Smith AC, Dykens E, Greenberg F. Smith AC, et al. Am J Med Genet. 1998 Mar 28;81(2):179-85. doi: 10.1002/(sici)1096-8628(19980328)81:2<179::aid-ajmg10>3.0.co;2-e. Am J Med Genet. 1998. PMID: 9613859 Review.
Cited by
- A statistical model to identify hereditary and epigenetic fusion genes associated with dilated cardiomyopathy.
Fei L, Zhang J, Zhuo D. Fei L, et al. Front Genet. 2024 Oct 1;15:1438887. doi: 10.3389/fgene.2024.1438887. eCollection 2024. Front Genet. 2024. PMID: 39411373 Free PMC article. - Novel crossover and recombination hotspots massively spread across primate genomes.
Ohadi M, Arabfard M, Khamse S, Alizadeh S, Vafadar S, Bayat H, Tajeddin N, Maddi AMA, Delbari A, Khorram Khorshid HR. Ohadi M, et al. Biol Direct. 2024 Aug 21;19(1):70. doi: 10.1186/s13062-024-00508-8. Biol Direct. 2024. PMID: 39169390 Free PMC article. - Copy number variations of chromosome 17p11.2 region in children with development delay and in fetuses with abnormal imaging findings.
Zhang Y, Liu X, Gao H, He R, Chu G, Zhao Y. Zhang Y, et al. BMC Med Genomics. 2021 Sep 1;14(1):215. doi: 10.1186/s12920-021-01065-z. BMC Med Genomics. 2021. PMID: 34470638 Free PMC article. - Prognostic significance of isochromosome 17q in hematologic malignancies.
Koczkodaj D, Muzyka-Kasietczuk J, Chocholska S, Podhorecka M. Koczkodaj D, et al. Oncotarget. 2021 Mar 30;12(7):708-718. doi: 10.18632/oncotarget.27914. eCollection 2021 Mar 30. Oncotarget. 2021. PMID: 33868591 Free PMC article. - 2018 Victor A. McKusick Leadership Award: Molecular Mechanisms for Genomic and Chromosomal Rearrangements.
Lupski JR. Lupski JR. Am J Hum Genet. 2019 Mar 7;104(3):391-406. doi: 10.1016/j.ajhg.2018.12.018. Am J Hum Genet. 2019. PMID: 30849326 Free PMC article. No abstract available.
References
Electronic-Database Information
- BLAST 2 Sequences, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html (for pairwise sequence comparison)
- Dotter program, http://www.cgr.ki.se/cgr/groups/sonnhammer/Dotter.html (for sequence comparison)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the distal SMS-REP [accession number BK001589], the middle SMS-REP [accession number BK001590], and the proximal SMS-REP [accession number BK001591])
- MFold program, http://www.biology.wustl.edu/gcg/mfold.html (for DNA secondary structure prediction)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMS) - PubMed
References
- Barbouti A, Stankiewicz P, Birren B, Nusbaum C, Cuomo C, Cook A, Höglund M, Johansson B, Hagemeijer A, Park S-S, Mitelman F, Lupski JR, Fioretos T. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large palindromic low-copy repeats. Am J Hum Genet (in press) - PMC - PubMed
- Bi W, Yan J, Stankiewicz P, Park S-S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJM, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–72810.1101/gr.73702 - DOI - PMC - PubMed
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HD2406407/HD/NICHD NIH HHS/United States
- M01 RR00188/RR/NCRR NIH HHS/United States
- M01 RR000188/RR/NCRR NIH HHS/United States
- P01 HD38420/HD/NICHD NIH HHS/United States
- R01 HD038420/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous