FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa - PubMed (original) (raw)
FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa
Bixing Huang et al. J Bacteriol. 2003 Dec.
Abstract
Twitching motility is a form of surface translocation mediated by the extension, tethering, and retraction of type IV pili. Three independent Tn5-B21 mutations of Pseudomonas aeruginosa with reduced twitching motility were identified in a new locus which encodes a predicted protein of unknown function annotated PA4959 in the P. aeruginosa genome sequence. Complementation of these mutants with the wild-type PA4959 gene, which we designated fimX, restored normal twitching motility. fimX mutants were found to express normal levels of pilin and remained sensitive to pilus-specific bacteriophages, but they exhibited very low levels of surface pili, suggesting that normal pilus function was impaired. The fimX gene product has a molecular weight of 76,000 and contains four predicted domains that are commonly found in signal transduction proteins: a putative response regulator (CheY-like) domain, a PAS-PAC domain (commonly involved in environmental sensing), and DUF1 (or GGDEF) and DUF2 (or EAL) domains, which are thought to be involved in cyclic di-GMP metabolism. Red fluorescent protein fusion experiments showed that FimX is located at one pole of the cell via sequences adjacent to its CheY-like domain. Twitching motility in fimX mutants was found to respond relatively normally to a range of environmental factors but could not be stimulated by tryptone and mucin. These data suggest that fimX is involved in the regulation of twitching motility in response to environmental cues.
Figures
FIG. 1.
Schematic representation of the fimX locus. The relevant restriction sites are indicated. (A) Overall topography of the fimX region. The open arrows indicate the relative transcriptional orientations of fimX and its neighboring genes, and Tt indicates the predicted transcription terminator following fimX. (B) Expanded view of fimX. The rectangles indicate the predicted domains in FimX. The transposon insertion sites in fimX mutants S19, S46, and S58 are indicated by solid triangles. The orientations of fimX in derived plasmid constructs are indicated by arrows; the arrows on the left indicate that the fimX coding region is in the same orientation as the adjacent lac promoter, and the arrows on the right indicate that the fimX coding region is in the same orientation as the adjacent T7 promoter. Note that the _Not_I and last _Sal_I restriction sites were derived from the multiple cloning site of the pUCPKS and pUCPSK vectors, which were used to construct translational fusions with RFP for subcellular localization studies.
FIG. 2.
Macroscopic and microscopic examination of twitching motility in fimX mutants. (A to F) Twitching zones observed in the subsurface stab assay on agar plates after 24 h of growth. Bar = 1 cm. (G to L) Light microscopy of the edges of the twitching zones at the interstitial surfaces between the glass coverslips and GelGro medium. Bar = 10 μm. (A and G) Wild-type strain PAK; (B and H) PAKΔ_pilA_ mutant; (C and I) fimX mutant S19; (D and J) S19(pUCPSK); (E and K) S19(pBH51); (F and L) S19(pBH52). The medium used for the subsurface twitching assay in complementation studies with the control vector or vectors containing fimX sequences contained 250 μg of carbenicillin ml−1. Similar results were obtained for complementation of mutants S46 and S58 (data not shown).
FIG. 3.
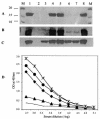
Western and ELISA analyses of pilus production in fimX mutants. (A) Surface pili extracted from PAK (lane 1), PAKΔ_pilA_ (lane 2), PAKΔ_pilV_ (lane 3), PAK(pUCPSK) (lane 4), PAK(pBH52) (lane 5), S19 (lane 6), S19(pUCPSK) (lane 7), and S19(pBH52) (lane 8). The gel was stained with Coomassie brilliant blue R250. (B) Western blotting of the surface pili from the same strains that were used in panel A. (C) Western blotting of the whole-cell proteins from the same strains that were used in panel A. (D) Quantitative analysis of the level of surface pili by ELISA for PAK (♦), PAKΔ_pilA_ (solid line), S19 (▴), S19(pBH51) (•), and S19(pBH52) (*). OD, optical density.
FIG. 4.
Comparison of the CheY-like, DUF1, and DUF2 domains of FimX with the domains of other proteins. (A) Alignment of the CheY-like domain of FimX with domains of other typical CheY-containing proteins. The five conserved functional sites identified by Volz (51) are indicated by asterisks. The sources of the CheY sequences (with their accession numbers) are as follows: CheY_Mdeg2095, putative CheY-like domain of Mdeg2095 in M. degradans; CheY_XAC2398, putative CheY-like domain of XAC2398 in X. axonopodis; CheY_Pa, CheY of P. aeruginosa (AAG04845); CheY_Ec, CheY of E. coli (NP_416396). (B and C) Alignment of the DUF1 and DUF2 domains of FimX with the corresponding domains of PleD, PdeA1 to PdeA3 and Dgc1 to Dgc3. The conserved functional sites in DUF1 and DUF2 are indicated by asterisks. The sequences are from references and .
FIG. 5.
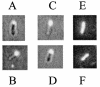
Localization of RFP fused with full-length FimX and with the CheY-like domain of FimX in PAK and S19. (A) PAK(pBH223) (RFP fused to FimX); (B) S19(pBH223); (C) PAK(pBH250) (RFP fused to the N-terminal region of FimX containing the CheY-like domain and adjacent sequences); (D) S19(pBH250); (E) PAK(pBH210) (RFP control); (F) S19(pBH210). Panels A to D were photographed with a dual light source to reveal the location of fluorescence in relation to the cell as a whole. The background is reduced in panels E and F, as only red fluorescent light and no background bright-field light were used in these cases.
FIG. 6.
Alignment of amino acid sequence adjacent to the CheY-like domain of FimX with amino acid sequences implicated in polar localization in other bacteria. The amino acid sequence between the _Sal_I site and the end of the CheY-like domain of FimX (SalI_FimX) was compared to the equivalent regions of other proteins implicated in polar localization in P. aeruginosa and other species, including PilS in P. aeruginosa (PilS_Pa) (20), MinD in N. gonorrhoeae (MinD_Ng) (36), CheZ in E. coli (CheZ_Ec) (10), and DivJ in C. crescentus (DivJ_Cc) (40).
Similar articles
- Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa.
Kazmierczak BI, Lebron MB, Murray TS. Kazmierczak BI, et al. Mol Microbiol. 2006 May;60(4):1026-43. doi: 10.1111/j.1365-2958.2006.05156.x. Mol Microbiol. 2006. PMID: 16677312 Free PMC article. - Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations.
Jain R, Behrens AJ, Kaever V, Kazmierczak BI. Jain R, et al. J Bacteriol. 2012 Aug;194(16):4285-94. doi: 10.1128/JB.00803-12. Epub 2012 Jun 8. J Bacteriol. 2012. PMID: 22685276 Free PMC article. - Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system--a review.
Darzins A, Russell MA. Darzins A, et al. Gene. 1997 Jun 11;192(1):109-15. doi: 10.1016/s0378-1119(97)00037-1. Gene. 1997. PMID: 9224880 Review. - Type IV pili and twitching motility.
Mattick JS. Mattick JS. Annu Rev Microbiol. 2002;56:289-314. doi: 10.1146/annurev.micro.56.012302.160938. Epub 2002 Jan 30. Annu Rev Microbiol. 2002. PMID: 12142488 Review.
Cited by
- A Cyclic di-GMP-binding Adaptor Protein Interacts with Histidine Kinase to Regulate Two-component Signaling.
Xu L, Venkataramani P, Ding Y, Liu Y, Deng Y, Yong GL, Xin L, Ye R, Zhang L, Yang L, Liang ZX. Xu L, et al. J Biol Chem. 2016 Jul 29;291(31):16112-23. doi: 10.1074/jbc.M116.730887. Epub 2016 May 26. J Biol Chem. 2016. PMID: 27231351 Free PMC article. - Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile.
Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Purcell EB, et al. J Bacteriol. 2012 Jul;194(13):3307-16. doi: 10.1128/JB.00100-12. Epub 2012 Apr 20. J Bacteriol. 2012. PMID: 22522894 Free PMC article. - Genetic Dissection of the Regulatory Network Associated with High c-di-GMP Levels in Pseudomonas putida KT2440.
Ramos-González MI, Travieso ML, Soriano MI, Matilla MA, Huertas-Rosales Ó, Barrientos-Moreno L, Tagua VG, Espinosa-Urgel M. Ramos-González MI, et al. Front Microbiol. 2016 Jul 20;7:1093. doi: 10.3389/fmicb.2016.01093. eCollection 2016. Front Microbiol. 2016. PMID: 27489550 Free PMC article. - A novel tetrameric PilZ domain structure from xanthomonads.
Li TN, Chin KH, Fung KM, Yang MT, Wang AH, Chou SH. Li TN, et al. PLoS One. 2011;6(7):e22036. doi: 10.1371/journal.pone.0022036. Epub 2011 Jul 7. PLoS One. 2011. PMID: 21760949 Free PMC article. - Role of cyclic Di-GMP during el tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic Di-GMP phosphodiesterase CdpA.
Tamayo R, Schild S, Pratt JT, Camilli A. Tamayo R, et al. Infect Immun. 2008 Apr;76(4):1617-27. doi: 10.1128/IAI.01337-07. Epub 2008 Jan 28. Infect Immun. 2008. PMID: 18227161 Free PMC article.
References
- Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. - PubMed
- Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. - PubMed
- Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources