FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism - PubMed (original) (raw)
FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism
E-H Lee et al. J Bacteriol. 2003 Dec.
Abstract
The farAB operon of Neisseria gonorrhoeae encodes an efflux pump which mediates gonococcal resistance to antibacterial fatty acids. It was previously observed that expression of the farAB operon was positively regulated by MtrR, which is a repressor of the mtrCDE-encoded efflux pump system (E.-H. Lee and W. M. Shafer, Mol. Microbiol. 33:839-845, 1999). This regulation was believed to be indirect since MtrR did not bind to the farAB promoter. In this study, computer analysis of the gonococcal genome sequence database, lacZ reporter fusions, and gel mobility shift assays were used to elucidate the regulatory mechanism by which expression of the farAB operon is modulated by MtrR in gonococci. We identified a regulatory protein belonging to the MarR family of transcriptional repressors and found that it negatively controls expression of farAB by directly binding to the farAB promoter. We designated this regulator FarR to signify its role in regulating the farAB operon. We found that MtrR binds to the farR promoter, thereby repressing farR expression. Hence, MtrR regulates farAB in a positive fashion by modulating farR expression. This MtrR regulatory cascade seems to play an important role in adjusting levels of the FarAB and MtrCDE efflux pumps to prevent their excess expression in gonococci.
Figures
FIG. 1.
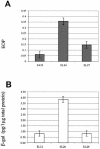
Effect of the marR1 mutation on FA resistance and farAB expression in N. gonorrhoeae FA19. (A) An EOP experiment was performed with strains FA19, EL24 (same as FA19 except marR1::Kmr) and EL27 (same as FA19 except marR2::Kmr) on GCB agar plates containing palmitic acid (150 μg/ml). EOPs are average values (± standard deviations [SD]) from at least three independent experiments. (B) Expression of farAB in EL12 (FA19[pLFAB1]) and its isogenic mutant strains EL26 (EL24[pLFAB1]) and EL29 (EL27[pLFAB1]). Shown are the amounts of β-Gal in cell extracts prepared as described in Materials and Methods from reporter strains EL12 and EL26, which contained the farAB::lacZ fusion. The results are averages of at least four independent experiments; error bars represent 1 SD.
FIG. 2.
Expression and purification of FarR-His. Protein samples collected during the purification were analyzed on SDS-15% PAGE gels stained with Coomassie brilliant blue. Lane 1, molecular weight standard markers (arrows [left], 17- and 32-kDa markers); lane 2, cell lysate after induction; lane 3, pooled fraction after Ni2+ affinity chromatography; lane 4, purified FarR after HPLC purification. Arrow (right), location of the FarR-His monomer.
FIG. 3.
DNA-binding properties of FarR. Shown is the binding of the purified His-tagged FarR protein to target DNA sequences. (A) FarR binding to farAB, farR, and mtrR-CDE promoter regions. Lanes 1, free labeled probe; lanes 2, probe with 0.2 ng of FarR; lanes 3, probe with 1 ng of FarR; lanes 4, probe with 5 ng of FarR. The probe used is indicated at the bottom of each panel. (B and C) Competition assays. (B) 32P-labeled 300-bp DNA encompassing the farAB promoter region was incubated with 5 ng of FarR. This binding was competed with unlabeled farAB (300 bp) or the mtrR-CDE intergenic region (310 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 0.1 μg of farAB DNA; lane 4, FarR with 1 μg of farAB DNA; lane 5, FarR with 0.1 μg of mtrR-CDE DNA; lane 6, FarR with 1 μg of mtrR-CDE DNA. (C) The 32P-labeled 305-bp farR promoter region was incubated with 5 ng of FarR. This binding was competed with the unlabeled farR promoter (305 bp) or a DNA sequence containing the farB coding region (365 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 1 μg of farR DNA; lane 4, FarR with 1 μg of farB DNA.
FIG. 3.
DNA-binding properties of FarR. Shown is the binding of the purified His-tagged FarR protein to target DNA sequences. (A) FarR binding to farAB, farR, and mtrR-CDE promoter regions. Lanes 1, free labeled probe; lanes 2, probe with 0.2 ng of FarR; lanes 3, probe with 1 ng of FarR; lanes 4, probe with 5 ng of FarR. The probe used is indicated at the bottom of each panel. (B and C) Competition assays. (B) 32P-labeled 300-bp DNA encompassing the farAB promoter region was incubated with 5 ng of FarR. This binding was competed with unlabeled farAB (300 bp) or the mtrR-CDE intergenic region (310 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 0.1 μg of farAB DNA; lane 4, FarR with 1 μg of farAB DNA; lane 5, FarR with 0.1 μg of mtrR-CDE DNA; lane 6, FarR with 1 μg of mtrR-CDE DNA. (C) The 32P-labeled 305-bp farR promoter region was incubated with 5 ng of FarR. This binding was competed with the unlabeled farR promoter (305 bp) or a DNA sequence containing the farB coding region (365 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 1 μg of farR DNA; lane 4, FarR with 1 μg of farB DNA.
FIG. 3.
DNA-binding properties of FarR. Shown is the binding of the purified His-tagged FarR protein to target DNA sequences. (A) FarR binding to farAB, farR, and mtrR-CDE promoter regions. Lanes 1, free labeled probe; lanes 2, probe with 0.2 ng of FarR; lanes 3, probe with 1 ng of FarR; lanes 4, probe with 5 ng of FarR. The probe used is indicated at the bottom of each panel. (B and C) Competition assays. (B) 32P-labeled 300-bp DNA encompassing the farAB promoter region was incubated with 5 ng of FarR. This binding was competed with unlabeled farAB (300 bp) or the mtrR-CDE intergenic region (310 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 0.1 μg of farAB DNA; lane 4, FarR with 1 μg of farAB DNA; lane 5, FarR with 0.1 μg of mtrR-CDE DNA; lane 6, FarR with 1 μg of mtrR-CDE DNA. (C) The 32P-labeled 305-bp farR promoter region was incubated with 5 ng of FarR. This binding was competed with the unlabeled farR promoter (305 bp) or a DNA sequence containing the farB coding region (365 bp). Lane 1, no protein added; lane 2, FarR; lane 3, FarR with 1 μg of farR DNA; lane 4, FarR with 1 μg of farB DNA.
FIG. 4.
MtrR binds to the DNA sequence upstream of farR. Shown is the binding of MBP-MtrR to target DNA sequences, the mtrR-CDE, farAB, and farR promoter regions. Lanes 1 (from left), free labeled probe; lanes 2, probe with 1.6 μg of MBP-MtrR; lanes 3, probe with 4 μg of MBP-MtrR. The probe used is indicated at the bottom.
FIG. 5.
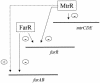
A model for MtrR regulation of farAB and mtrCDE efflux pump operons in N. gonorrhoeae. This model describes the ability of MtrR to positively regulate (+) farAB expression by repressing (−) farR and mtrCDE expression. This MtrR regulatory circuit is most likely to be important in preventing the excess expression of these efflux pumps in gonococci.
Similar articles
- Integration Host Factor is required for FarR repression of the farAB-encoded efflux pump of Neisseria gonorrhoeae.
Lee EH, Hill SA, Napier R, Shafer WM. Lee EH, et al. Mol Microbiol. 2006 Jun;60(6):1381-400. doi: 10.1111/j.1365-2958.2006.05185.x. Mol Microbiol. 2006. PMID: 16796676 - Dueling regulatory properties of a transcriptional activator (MtrA) and repressor (MtrR) that control efflux pump gene expression in Neisseria gonorrhoeae.
Zalucki YM, Dhulipala V, Shafer WM. Zalucki YM, et al. mBio. 2012 Dec 4;3(6):e00446-12. doi: 10.1128/mBio.00446-12. mBio. 2012. PMID: 23221802 Free PMC article. - The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids.
Lee EH, Shafer WM. Lee EH, et al. Mol Microbiol. 1999 Aug;33(4):839-45. doi: 10.1046/j.1365-2958.1999.01530.x. Mol Microbiol. 1999. PMID: 10447892 - Transcriptional regulation of the mtrCDE efflux pump operon: importance for Neisseria gonorrhoeae antimicrobial resistance.
Ayala JC, Balthazar JT, Shafer WM. Ayala JC, et al. Microbiology (Reading). 2022 Aug;168(8):001231. doi: 10.1099/mic.0.001231. Microbiology (Reading). 2022. PMID: 35916832 Free PMC article. Review. - Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis.
Shafer WM, Veal WL, Lee EH, Zarantonelli L, Balthazar JT, Rouquette C. Shafer WM, et al. J Mol Microbiol Biotechnol. 2001 Apr;3(2):219-24. J Mol Microbiol Biotechnol. 2001. PMID: 11321577 Review.
Cited by
- Thiazoplanomicin, a new thiazolyl peptide antibiotic from the leaf-litter actinomycete Actinoplanes sp. MM794L-181F6.
Takehana Y, Muramatsu H, Hatano M, Ishizaki Y, Umekita M, Shibuya Y, Hayashi C, Kimura T, Takeuchi T, Shimuta K, Sawa R, Igarashi M. Takehana Y, et al. J Antibiot (Tokyo). 2024 Oct 28. doi: 10.1038/s41429-024-00783-7. Online ahead of print. J Antibiot (Tokyo). 2024. PMID: 39468287 - Regulation Mechanism of Nicotine Catabolism in Sphingomonas melonis TY by a Dual Role Transcriptional Regulator NdpR.
Wang H, Wang X, Tang Q, Wang L, Mei C, Shao Y, Xu Y, Lu Z, Zhong W. Wang H, et al. Appl Environ Microbiol. 2023 May 31;89(5):e0032423. doi: 10.1128/aem.00324-23. Epub 2023 Apr 18. Appl Environ Microbiol. 2023. PMID: 37071026 Free PMC article. - The Neisseria gonorrhoeae type IV pilus promotes resistance to hydrogen peroxide- and LL-37-mediated killing by modulating the availability of intracellular, labile iron.
Hu LI, Stohl EA, Seifert HS. Hu LI, et al. PLoS Pathog. 2022 Jun 17;18(6):e1010561. doi: 10.1371/journal.ppat.1010561. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35714158 Free PMC article. - Neisseria genes required for persistence identified via in vivo screening of a transposon mutant library.
Rhodes KA, Ma MC, Rendón MA, So M. Rhodes KA, et al. PLoS Pathog. 2022 May 17;18(5):e1010497. doi: 10.1371/journal.ppat.1010497. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35580146 Free PMC article. - Joint engineering of SACE_Lrp and its target MarR enhances the biosynthesis and export of erythromycin in Saccharopolyspora erythraea.
Liu J, Li L, Wang Y, Li B, Cai X, Tang L, Dong S, Yang E, Wu H, Zhang B. Liu J, et al. Appl Microbiol Biotechnol. 2021 Apr;105(7):2911-2924. doi: 10.1007/s00253-021-11228-8. Epub 2021 Mar 24. Appl Microbiol Biotechnol. 2021. PMID: 33760930
References
- Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. - PubMed
- Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. - PubMed
- Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. - PubMed
- Delahay, R. M., B. D. Robertson, J. T. Balthazar, W. M. Shafer, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic compounds. Microbiology 143:2127-2133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI021150/AI/NIAID NIH HHS/United States
- RR13948/RR/NCRR NIH HHS/United States
- 5T32 AI-07470/AI/NIAID NIH HHS/United States
- AI-38399/AI/NIAID NIH HHS/United States
- R37 AI021150/AI/NIAID NIH HHS/United States
- RR12878/RR/NCRR NIH HHS/United States
- AI-37945/AI/NIAID NIH HHS/United States
- AI-21150-17/AI/NIAID NIH HHS/United States
- RR02878/RR/NCRR NIH HHS/United States
- P01 AI037945/AI/NIAID NIH HHS/United States
- T32 AI007470/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources