Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5' untranslated regions - PubMed (original) (raw)
Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5' untranslated regions
Sergei E Dmitriev et al. Mol Cell Biol. 2003 Dec.
Abstract
The reconstitution of translation initiation complexes from purified components is a reliable approach to determine the complete set of essential canonical initiation factors and auxiliary proteins required for the 40S ribosomal subunit to locate the initiation codon on individual mRNAs. Until now, it has been successful mostly for formation of 48S translation initiation complexes with viral IRES elements. Among cap-dependent mRNAs, only globin mRNAs and transcripts with artificial 5' leaders were amenable to this assembly. Here, with modified conditions for the reconstitution, 48S complexes have been successfully assembled with the 5' UTR of beta-actin mRNA (84 nucleotides) and the tripartite leader of adenovirus RNAs (232 nucleotides), though the latter has been able to use only the scanning rather then the shunting model of translation initiation with canonical initiation factors. We show that initiation factor 4B is essential for mRNAs that have even a rather moderate base pairing within their 5' UTRs (with the cumulative stability of the secondary structure within the entire 5' UTR < -13 kcal/mol) and not essential for beta-globin mRNA. A recombinant eIF4B poorly substitutes for the native factor. The 5' UTRs with base-paired G residues reveal a very sharp dependence on the eIF4B concentration to form the 48S complex. The data suggest that even small variations in concentration or activity of eIF4B in mammalian cells may differentially affect the translation of different classes of cap-dependent cellular mRNAs.
Figures
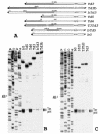
FIG. 1.
(A) A schematic representation of the tripartite leader (TPL) of adenovirus 2 and its deletion derivatives fused to beta-globin mRNA. Shaded and open bars represent the tripartite leader and beta-globin sequences, respectively. Figures without parentheses above the bars show the terminal nucleotides of the tripartite leader that are still present in the constructs. Those in parentheses denote nucleotides from the beta-globin sequence. (B) Primer extension analysis (toeprinting) of 48S initiation complexes assembled with the transcripts shown in A. Lane control corresponds to the complex reconstituted in the absence of Met-tRNAi and eIF2. The reconstitution was performed with total tRNA (where only tRNAiMet was aminoacylated) and recombinant eIF4B. A dideoxynucleotide sequence of pTbG generated with the same primer was run in parallel. For other details see the text. (C) Toeprint analysis of 48S initiation complexes formation in rabbit reticulocyte lysate in the presence of GMP-PNP with the same set of transcripts. In lane control, the assembly was inhibited by addition of 30 mM Mg2+.
FIG. 2.
(A) A schematic representation of construct Td3 and its different modifications. Boxed TTT in construct T3Td3 denote a substitution of three consecutive G residues in construct Td3 for three T residues. Other designations are as in Fig. 1A. (B) Toeprint analysis of 48S initiation complexes assembled on mRNAs bG, Td3, Td5, Td6, bTd3, and Td3b. A dideoxynucleotide sequence generated with the same primer corresponds to construct Td3. (C) Toeprint analysis of 48S initiation complexes reconstituted with model mRNAs bTd5 and T3Td3. For comparison, the complexes assembled on Td3 and bG were analyzed in parallel. A dideoxynucleotide sequence generated with the same primer corresponds to construct Td3.
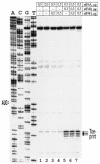
FIG. 3.
Accessibility of nucleotide residues in the 5′ termini of constructs bG, Td3, Td5, Td6, and T3Td3 to chemical and enzymatic attack (for details see 6). The sequence corresponding to the beta-globin mRNA is shown in bold. In our hands, the C- and U-tracts are poorly attacked by RNases and chemicals, and hence their secondary structure status is not clear.
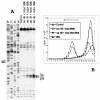
FIG. 4.
(A) Toeprint analysis of 48S initiation complexes assembled on mRNA Td3 with various combinations of eIF4B and Met-tRNAi preparations. rec 4B and nat 4B, recombinant and native eIF4B, respectively. Total tRNA corresponds to the total calf liver tRNA (Novagen), where only tRNAiMet was amynoacylated; indiv. tRNA corresponds to individual Met-tRNAi. A dideoxynucleotide sequence of pTd3 generated with the same primer is shown to the left. (B) Sucrose gradient sedimentation of 48S complexes assembled under different conditions on mRNA Td3. The mRNA was capped and labeled by incorporation of [32P]UTP by T7 transcription reaction. Curve control corresponds to the complex reconstituted in the absence of Met-tRNAiMet and eIF2. The direction of sedimentation is shown by the arrow.
FIG. 5.
(A) Toeprinting of 48S complexes assembled on mRNAs Td3, bG, and Td6 with addition of different amounts of native eIF4B. The amounts of eIF4B per standard volume of the reconstitution mixture (20 μl) were as follows: lanes 2 and 7, 11 to 0.025 μg, lanes 3 and 8, 12 to 0.05 μg, lanes 4 and 9, 13 to 0.1 μg, lanes 5 and 10, 14 to 0.2 μg, and lane 6, 0.3 μg. The yields of 48S complexes for Td6 and bG with 0.3 μg of eIF4B were the same as with 0.2 μg of eIF4B, and the respective lanes are not presented in the figure. No exogenous eIF4B was added to the samples corresponding to lanes 2, 7, and 11. The indicated amount of eIF4B in these samples is accounted for by contamination of affinity purified eIF4F with eIF4B. It was estimated by quantitative Western blotting as 0.05 μg of eIF4B/1 μg of eIF4F. Lane bG control corresponds to the 48S complex reconstituted with mRNA bG in the absence of Met-tRNAiMet and eIF2. A dideoxynucleotide sequence was generated from pTd3. (B) Dependence of formation of 48S complexes assembled on mRNAs bG, Td3, and Td6 on the molar concentration of eIF4B in the reconstitution mixture. The yield of 48S complexes (radioactivity in toeprint bands) was determined with phosphorimager from data presented in A and presented as the ratio to the maximal yield of the complexes for each mRNA.
FIG. 6.
Effect of substitution of eIF4B for eIF4H on the reconstitution of 48S complexes with mRNA Td3. Recombinant and native eIF4H gave similar results. The data in the figure were obtained with native eIF4H. Amounts of factors eIF4A, eIF4B, and eIF4H indicated above the lanes are given per standard volume of the reconstitution mixture (20 μl). A dideoxynucleotide sequence of pTd3 is presented to the left.
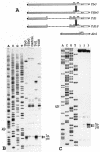
FIG. 7.
(A) A schematic representation of constructs TbG, Td1, its derivatives with a low-energy stem-loop structures inserted into position 204 of the tripartite leader, and construct AbG. Shaded bars represent the tripartite leader or beta-actin sequences. Other designations are as in Fig. 1A. (B) Reconstitution of the 48S complex with the mRNAs TbG, Td1, TSbG, and Td1S. The individual in vitro-transcribed tRNAiMet and native eIF4B were used in this experiment. Control shows the assembly in the absence of Met-tRNAiMet and eIF2. A dideoxynucleotide sequence was obtained from pTbG. (C) Assembly of the 48S complex with the mRNA carrying the 5′ UTR of human beta-actin mRNA. 1, control (no Met-tRNAiMet, no eIF2); 2, reconstitution with the total tRNA and recombinant eIF4B; 3, reconstitution with in vitro-transcribed individual Met-tRNAiMet and native eIF4B (0.5 μg/20 μl). A dideoxynucleotide sequence generated from pAbG with the same primer was run in parallel.
Similar articles
- Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting.
Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN. Dmitriev SE, et al. FEBS Lett. 2003 Jan 2;533(1-3):99-104. doi: 10.1016/s0014-5793(02)03776-6. FEBS Lett. 2003. PMID: 12505166 - The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection.
Pestova TV, Kolupaeva VG. Pestova TV, et al. Genes Dev. 2002 Nov 15;16(22):2906-22. doi: 10.1101/gad.1020902. Genes Dev. 2002. PMID: 12435632 Free PMC article. - [Minor secondary-structure variation in the 5'-untranslated region of the beta-globin mRNA changes the concentration requirements for eIF2].
Dmitriev SE, Terenin IM, Rubtsova MP, Shatskiĭ IN. Dmitriev SE, et al. Mol Biol (Mosk). 2003 May-Jun;37(3):494-503. Mol Biol (Mosk). 2003. PMID: 12815957 Russian. - mRNA helicases: the tacticians of translational control.
Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. Parsyan A, et al. Nat Rev Mol Cell Biol. 2011 Apr;12(4):235-45. doi: 10.1038/nrm3083. Nat Rev Mol Cell Biol. 2011. PMID: 21427765 Review. - The role of the 5' untranslated region of an mRNA in translation regulation during development.
van der Velden AW, Thomas AA. van der Velden AW, et al. Int J Biochem Cell Biol. 1999 Jan;31(1):87-106. doi: 10.1016/s1357-2725(98)00134-4. Int J Biochem Cell Biol. 1999. PMID: 10216946 Review.
Cited by
- Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis.
Terenin IM, Akulich KA, Andreev DE, Polyanskaya SA, Shatsky IN, Dmitriev SE. Terenin IM, et al. Nucleic Acids Res. 2016 Feb 29;44(4):1882-93. doi: 10.1093/nar/gkv1514. Epub 2015 Dec 29. Nucleic Acids Res. 2016. PMID: 26717981 Free PMC article. - Regulatory BC1 RNA in cognitive control.
Iacoangeli A, Dosunmu A, Eom T, Stefanov DG, Tiedge H. Iacoangeli A, et al. Learn Mem. 2017 Jun 15;24(7):267-277. doi: 10.1101/lm.045427.117. Print 2017 Jul. Learn Mem. 2017. PMID: 28620074 Free PMC article. - MMP13 inhibition rescues cognitive decline in Alzheimer transgenic mice via BACE1 regulation.
Zhu BL, Long Y, Luo W, Yan Z, Lai YJ, Zhao LG, Zhou WH, Wang YJ, Shen LL, Liu L, Deng XJ, Wang XF, Sun F, Chen GJ. Zhu BL, et al. Brain. 2019 Jan 1;142(1):176-192. doi: 10.1093/brain/awy305. Brain. 2019. PMID: 30596903 Free PMC article. - Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases.
Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Raught B, et al. EMBO J. 2004 Apr 21;23(8):1761-9. doi: 10.1038/sj.emboj.7600193. Epub 2004 Apr 8. EMBO J. 2004. PMID: 15071500 Free PMC article. - Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting.
Xi Q, Cuesta R, Schneider RJ. Xi Q, et al. Genes Dev. 2004 Aug 15;18(16):1997-2009. doi: 10.1101/gad.1212504. Genes Dev. 2004. PMID: 15314025 Free PMC article.
References
- Bi, X., Ren, J., and D. J. Goss, 2000. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39:5758-5765. - PubMed
- Browning, K. S., S. R. Lax, J. Humphreys, J. M. Ravel, S. A. Jobling, and L. Gehrke. 1988. Evidence that the 5′-untranslated leader of mRNA affects the requirement for wheat germ initiation factors 4A, 4F, and 4G (4B). J. Biol. Chem. 263:9630-9634. - PubMed
- Dmitriev, S. E., A. V. Pisarev, M. P. Rubtsova, Y. E. Dunaevsky, and I. N. Shatsky. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 533:99-104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous