Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster - PubMed (original) (raw)
Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster
Mathieu Miron et al. Mol Cell Biol. 2003 Dec.
Abstract
The eIF4E-binding proteins (4E-BPs) interact with translation initiation factor 4E to inhibit translation. Their binding to eIF4E is reversed by phosphorylation of several key Ser/Thr residues. In Drosophila, S6 kinase (dS6K) and a single 4E-BP (d4E-BP) are phosphorylated via the insulin and target of rapamycin (TOR) signaling pathways. Although S6K phosphorylation is independent of phosphoinositide 3-OH kinase (PI3K) and serine/threonine protein kinase Akt, that of 4E-BP is dependent on PI3K and Akt. This difference prompted us to examine the regulation of d4E-BP in greater detail. Analysis of d4E-BP phosphorylation using site-directed mutagenesis and isoelectric focusing-sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated that the regulatory interplay between Thr37 and Thr46 of d4E-BP is conserved in flies and that phosphorylation of Thr46 is the major phosphorylation event that regulates d4E-BP activity. We used RNA interference (RNAi) to target components of the PI3K, Akt, and TOR pathways. RNAi experiments directed at components of the insulin and TOR signaling cascades show that d4E-BP is phosphorylated in a PI3K- and Akt-dependent manner. Surprisingly, RNAi of dAkt also affected insulin-stimulated phosphorylation of dS6K, indicating that dAkt may also play a role in dS6K phosphorylation.
Figures
FIG. 1.
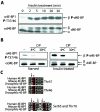
Insulin treatment of S2 cells and potential phosphorylation sites of d4E-BP. (A) Time course of d4E-BP phosphorylation in response to insulin. S2 cells were starved for 36 h and stimulated with bovine insulin (1 μg/ml) for the indicated times. (B) Phosphatase treatment. Extracts from starved or insulin-treated S2 cells were treated with CIP for 30 min at the indicated temperatures. The effect of insulin treatment on d4E-BP in panels A and B was assessed by immunoblotting with anti-phospho-4E-BP1(Thr37/46) (α4E-BP1) (top) and by gel shift of the α phosphoisoform to β as visualized by anti-d4E-BP (αd4E-BP) (bottom) using extracts from S2 cells. P, phosphorylated. (C) Sequence alignment of the putative phosphorylation sites of d4E-BP and human and murine 4E-BP1. Identical (solid boxes) and conserved (shaded boxes) amino acids are highlighted. Conserved phosphorylation sites are colored red and marked by asterisks. The arrows indicate differences in (for Thr37, Ser65, and Thr70) or identity of (for Thr46) the amino acids surrounding the putative phosphorylation sites of d4E-BP.
FIG. 2.
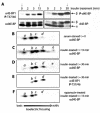
Analysis of mutations at d4E-BP phosphorylation sites. (A) Mutation of Thr37 and/or Thr46 to Ala (T37A and T46A, respectively) prevents the insulin-induced phosphorylation of d4E-BP. As expected, the double mutant (T37A/T46A) also showed no phosphorylation. The last lane (-) is an untransfected control. WT, wild type; +, treated; −, untreated; P, phosphorylated. (B) Mutation of Ser65 and/or Thr70 to Ala (S65A and T70A, respectively) had no effect on phosphorylation. As expected, the double mutant (S65A/T70A) also had no effect. S2 cells transfected with 3HA-tagged d4E-BP mutant cDNAs were starved for 24 h before insulin treatment. The effect of insulin was assessed with anti-phospho-4E-BP1(Thr37/46) (α4E-BP1 P-T37/46) (middle), and protein expression levels were compared using anti-HA-11 (αHA) (bottom). Total protein loading levels were compared using anti-deIF4A (αdeIF4A).
FIG. 3.
Analysis by IEF-SDS-PAGE of d4E-BP phosphorylation. (A) Selected S2 cell extracts for IEF-SDS-PAGE (arrows). Rapamycin-treated cell extract (not shown) is similar to unstimulated cell extract. (B to F) IEF-SDS-PAGE of extracts from serum-starved (B), insulin-treated (15 min) (C), insulin-treated (30 min) (D and E), and rapamycin- and insulin-treated (15 min) (F) cells were probed with the indicated antibodies. d4E-BP (B to F) was assessed by blotting it with anti-d4E-BP (αd4E-BP) or anti-phospho-4E-BP1(Thr37/46) (α4E-BP1 P-T37/46) as indicated. The individual isoforms are represented by the letters a to e. P, phosphorylated.
FIG. 4.
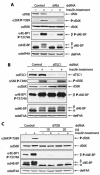
Effects of RNAi of dPI3K, dPTEN, and dPDK1 on d4E-BP phosphorylation in S2 cells. (A) Northern analysis confirms that the dPI3K (Dp110) and dPTEN transcript amounts are reduced compared with control cells. (B) RNAis of dPI3K and dPTEN have different effects on d4E-BP phosphorylation. Whereas dPI3K RNAi reduces the insulin-stimulated phosphorylation of d4E-BP, dPTEN RNAi increases the phosphorylation of d4E-BP in unstimulated cells. The phosphorylation of dS6K Thr398 is increased by RNAi of dPI3K but not by RNAi of dPTEN. RNAi of Dp110 and dPTEN was performed at least three times. +, treated; −, untreated. (C) Northern analysis confirms that the dPDK1 transcript is reduced compared with control cells. (D) A reduction in dPDK1 amounts reduces the endogenous levels of d4E-BP but does not affect its insulin-induced phosphorylation. dPDK1 RNAi does, however, block phosphorylation of dS6K at Thr398. RNAi of dPDK1 was performed four times. The effect on d4E-BP was assessed by blotting with anti-phospho-4E-BP1(Thr37/46) (α4E-BP1 P-T37/46) and by gel shift of the α phosphoisoform to β with anti-d4E-BP (αd4E-BP). The phosphorylation of dS6K was detected with anti-phospho-S6K Thr389 (αS6K P-T389). Antibodies against translation factors deIF4E (αdeIF4E) and deIF4A (αdeIF4A) were used to confirm equal loading of the gels in panels B and D, respectively. RpS15a is the probe for ribosomal protein S15a, which was used to confirm equal loading in panels A and C. P, phosphorylated.
FIG. 5.
Effects of RNAi of dAkt, dTSC1, and dTOR on d4E-BP phosphorylation in S2 cells. (A) A reduction in the amount of dAkt blocks signaling to d4E-BP and reduces insulin-induced phosphorylation. The phosphorylation of dS6K at Thr398 is also reduced. +, treated; −, untreated; P, phosphorylated. (B) A reduction in the amount of dTsc1 augments signaling to d4E-BP, causing an increase in the basal levels of phosphorylated d4E-BP. The phosphorylation of dS6K at Thr398 is also increased. (C) A reduction in the amount of dTOR blocks signaling to d4E-BP and dS6K and abolishes the insulin-induced phosphorylation of both proteins. RNAis of dAkt, dTSC1, and dTOR were performed at least three times. The effects on d4E-BP and dS6K were assessed as for Fig. 4.
FIG. 6.
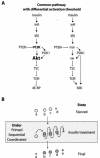
Models of the insulin-signaling pathway in Drosophila and regulation of d4E-BP by phosphorylation. (A) A common pathway regulates 4E-BP and S6K. The regulation of 4E-BP is dependent on signaling from Inr-PI3K-Akt-TSC and TOR. The regulation of S6K is also effected by PDK1 and the same upstream elements. The different size of the Akt typeface is to illustrate that the intensity of signaling toward TSC affects 4E-BP and S6K differentially. (B) The pattern of phosphorylation of d4E-BP as observed by IEF-SDS-PAGE may result from one of three possible models. (i) In the primed model, d4E-BP is already phosphorylated at Thr37 and is subsequently phosphorylated on one additional site, Thr46, after insulin stimulation. (ii) In the sequential model, d4E-BP is phosphorylated on Thr37 and then Thr46 (or vice versa). (iii) In the coordinated model, d4E-BP is phosphorylated coordinately on Thr37 and Thr46.
Similar articles
- Insulin-induced Drosophila S6 kinase activation requires phosphoinositide 3-kinase and protein kinase B.
Lizcano JM, Alrubaie S, Kieloch A, Deak M, Leevers SJ, Alessi DR. Lizcano JM, et al. Biochem J. 2003 Sep 1;374(Pt 2):297-306. doi: 10.1042/BJ20030577. Biochem J. 2003. PMID: 12841848 Free PMC article. - Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling.
Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Radimerski T, et al. Genes Dev. 2002 Oct 15;16(20):2627-32. doi: 10.1101/gad.239102. Genes Dev. 2002. PMID: 12381661 Free PMC article. - Pharmacogenomic profiling of the PI3K/PTEN-AKT-mTOR pathway in common human tumors.
Xu G, Zhang W, Bertram P, Zheng XF, McLeod H. Xu G, et al. Int J Oncol. 2004 Apr;24(4):893-900. Int J Oncol. 2004. PMID: 15010827 - Regulation of translation initiation by FRAP/mTOR.
Gingras AC, Raught B, Sonenberg N. Gingras AC, et al. Genes Dev. 2001 Apr 1;15(7):807-26. doi: 10.1101/gad.887201. Genes Dev. 2001. PMID: 11297505 Review. No abstract available. - A Role of PI3K/Akt Signaling in Oocyte Maturation and Early Embryo Development.
Kalous J, Aleshkina D, Anger M. Kalous J, et al. Cells. 2023 Jul 12;12(14):1830. doi: 10.3390/cells12141830. Cells. 2023. PMID: 37508495 Free PMC article. Review.
Cited by
- 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth.
Teleman AA, Chen YW, Cohen SM. Teleman AA, et al. Genes Dev. 2005 Aug 15;19(16):1844-8. doi: 10.1101/gad.341505. Genes Dev. 2005. PMID: 16103212 Free PMC article. - TOR signaling is required for amino acid stimulation of early trypsin protein synthesis in the midgut of Aedes aegypti mosquitoes.
Brandon MC, Pennington JE, Isoe J, Zamora J, Schillinger AS, Miesfeld RL. Brandon MC, et al. Insect Biochem Mol Biol. 2008 Oct;38(10):916-22. doi: 10.1016/j.ibmb.2008.07.003. Epub 2008 Jul 29. Insect Biochem Mol Biol. 2008. PMID: 18708143 Free PMC article. - Statins Induce Locomotion and Muscular Phenotypes in Drosophila melanogaster That Are Reminiscent of Human Myopathy: Evidence for the Role of the Chloride Channel Inhibition in the Muscular Phenotypes.
Al-Sabri MH, Behare N, Alsehli AM, Berkins S, Arora A, Antoniou E, Moysiadou EI, Anantha-Krishnan S, Cosmen PD, Vikner J, Moulin TC, Ammar N, Boukhatmi H, Clemensson LE, Rask-Andersen M, Mwinyi J, Williams MJ, Fredriksson R, Schiöth HB. Al-Sabri MH, et al. Cells. 2022 Nov 8;11(22):3528. doi: 10.3390/cells11223528. Cells. 2022. PMID: 36428957 Free PMC article. - Somatic stem cell differentiation is regulated by PI3K/Tor signaling in response to local cues.
Amoyel M, Hillion KH, Margolis SR, Bach EA. Amoyel M, et al. Development. 2016 Nov 1;143(21):3914-3925. doi: 10.1242/dev.139782. Epub 2016 Sep 15. Development. 2016. PMID: 27633989 Free PMC article. - IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback.
Marr MT 2nd, D'Alessio JA, Puig O, Tjian R. Marr MT 2nd, et al. Genes Dev. 2007 Jan 15;21(2):175-83. doi: 10.1101/gad.1506407. Genes Dev. 2007. PMID: 17234883 Free PMC article.
References
- Backman, S. A., V. Stambolic, A. Suzuki, J. Haight, A. Elia, J. Pretorius, M. S. Tsao, P. Shannon, B. Bolon, G. O. Ivy, and T. W. Mak. 2001. Deletion of PTEN in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29:396-403. - PubMed
- Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. - PubMed
- Britton, J. S., W. K. Lockwood, L. Li, S. M. Cohen, and B. A. Edgar. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2:239-249. - PubMed
- Brunn, G. J., P. Fadden, T. A. J. Haystead, and J. C. Lawrence, Jr. 1997. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J. Biol. Chem. 272:32547-32550. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous