Mutation of MEF2A in an inherited disorder with features of coronary artery disease - PubMed (original) (raw)
Mutation of MEF2A in an inherited disorder with features of coronary artery disease
Lejin Wang et al. Science. 2003.
Abstract
The early genetic pathway(s) triggering the pathogenesis of coronary artery disease (CAD) and myocardial infarction (MI) remain largely unknown. Here, we describe an autosomal dominant form of CAD/MI (adCAD1) that is caused by the deletion of seven amino acids in transcription factor MEF2A. The deletion disrupts nuclear localization of MEF2A, reduces MEF2A-mediated transcription activation, and abolishes synergistic activation by MEF2A and by the transcription factor GATA-1 through a dominant-negative mechanism. The MEF2A protein demonstrates strong expression in the endothelium of coronary arteries. These results identify a pathogenic gene for a familial vascular disease with features of CAD and implicate the MEF2A signaling pathway in the pathogenesis of CAD/MI.
Figures
Fig. 1
Genetic linkage of CAD/MI to chromosome 15q26 (adCAD1). (A) Pedigree structure and genotypic analysis of kindred QW1576. Individuals with CAD ( Table 1) are indicated by solid squares (males) or solid circles (females). Unaffected individuals are indicated by open symbols. Normal males under the age of 50 years or normal females under 55 years are shown in light-gray as uncertain phenotype. Deceased individuals are indicated by a slash (/). The proband is indicated by an arrow. Genotypes for markers D15S1014, D15S212, D15S120, and D15S87 are shown below each symbol. Initial linkage was identified with D15S120, which yielded a lod score of 4.19 at a recombination fraction of 0. Haplotype cosegregating with the disease is indicated by the black vertical bars. (B) Coronary angiogram from the proband, who experienced an inferior MI attributed to a plaque rupture lesion (arrow) with a 70% narrowing in the distal right coronary artery. This lesion is at a bifurcation site typical of the pattern of coronary atherosclerosis. The lesion was stented, and follow-up angiography of the site demonstrated wide patency, without any renarrowing. (C) Ideogram of chromosome 15 with the Geimsa banding pattern and localization of the adCAD1 locus. The genetic map with chromosome 15q26 markers and the location of the MEF2A gene is shown on the right.
Fig. 2
MEF2A intragenic deletion cosegregates with CAD in kindred QW1576. (A) The pedigree of kindred QW1576, showing genetic status: + indicates the presence of the 21-bp deletion of MEF2A (heterozygous); − indicates the absence of the deletion. (B) DNA sequence analysis of the wild-type ( WT ) allele and the 21-bp deletion allele (Δ21bp) of MEF2A. Sequence analysis of exon 11 of MEF2A in the proband (II.1) revealed the presence of a deletion. The wild-type and deletion alleles were separated by a 3% agarose gel and a single strand conformation polymorphism gel, purified and sequenced directly. The location of Δ21bp is indicated. (C) Δ21bp results in a deletion of seven amino acids of MEF2A (ΔQ440P441P442Q443P444Q445P446).
Fig. 3
Functional characterization of wild-type and Δ7aa MEF2A proteins by immunofluorescence. (A to C) MEF2A deletion Δ7aa causes a defect in nuclear localization of the MEF2A protein in three cell types: (A) human umbilical vascular endothelial cells (HUVEC); (B) human aortic smooth muscle cells (HVSMC); and (C) HeLa cells. Cells were transfected with expression constructs for wild-type and mutant MEF2A proteins tagged with a FLAG epitope. Green, MEF2A signal; blue, nucleus. DAPI, 4′,6-diamidino-2-phenylindole. (D) Colocalization of MEF2A and CD31 (PECAM, an endothelial cell–specific marker) in the endothelium of human coronary arteries. Cryosections (6 μm thick) of human coronary arteries were immunostained with the rabbit polyclonal antiserum to MEF2A. The adjacent sections were used for immunostaining with a monoclonal antibody to CD31. L, lumen; E, endothelium.
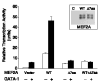
Fig. 4
Functional characterization of wild-type and Δ7aa MEF2A proteins by transcriptional activation assays. The effect of Δ7aa on the transcription activation activity of MEF2A was analyzed in the presence or absence of GATA-1 with the ANF_−700 promoter. Transcriptional activity is shown as relative luciferase activity on the y axis. The transcriptional activity of the reporter gene only (vector) was set arbitrarily to 1. WT/Δ7aa data represents coexpression of both wild-type and mutant MEF2As. Inset: Western blot analysis with rabbit polyclonal antiserum to MEF2A showed that both wild-type and mutant MEF2A were successfully expressed in transfected HeLa cells. Vector only (C) was used as a negative control. The MEF2A antibody detected two bands as previously reported (). The data shown are from two independent experiments in triplicate and are expressed as mean ± SEM._
## Similar articles
* Transcription factor MEF2A mutations in patients with coronary artery disease. Bhagavatula MR, Fan C, Shen GQ, Cassano J, Plow EF, Topol EJ, Wang Q. Bhagavatula MR, et al. Hum Mol Genet. 2004 Dec 15;13(24):3181-8. doi: 10.1093/hmg/ddh329. Epub 2004 Oct 20. Hum Mol Genet. 2004. PMID: 15496429 Free PMC article. * Advances in the genetic basis of coronary artery disease. Wang Q. Wang Q. Curr Atheroscler Rep. 2005 May;7(3):235-41. doi: 10.1007/s11883-005-0012-6. Curr Atheroscler Rep. 2005. PMID: 15811259 Free PMC article. Review. * Lack of MEF2A mutations in coronary artery disease. Weng L, Kavaslar N, Ustaszewska A, Doelle H, Schackwitz W, Hébert S, Cohen JC, McPherson R, Pennacchio LA. Weng L, et al. J Clin Invest. 2005 Apr;115(4):1016-20. doi: 10.1172/JCI24186. J Clin Invest. 2005. PMID: 15841183 Free PMC article. * MEF2A sequence variants and coronary artery disease: a change of heart? Altshuler D, Hirschhorn JN. Altshuler D, et al. J Clin Invest. 2005 Apr;115(4):831-3. doi: 10.1172/JCI24715. J Clin Invest. 2005. PMID: 15841171 Free PMC article. * Coronary artery disease and the MEF2A transcription factor. Olson EN. Olson EN. Sci Aging Knowledge Environ. 2003 Dec 3;2003(48):pe33. doi: 10.1126/sageke.2003.48.pe33. Sci Aging Knowledge Environ. 2003. PMID: 14657507 Review.
## Cited by
* Coronary artery disease susceptibility gene ADTRP regulates cell cycle progression, proliferation, and apoptosis by global gene expression regulation. Luo C, Wang F, Qin S, Chen Q, Wang QK. Luo C, et al. Physiol Genomics. 2016 Aug 1;48(8):554-64. doi: 10.1152/physiolgenomics.00028.2016. Epub 2016 May 27. Physiol Genomics. 2016. PMID: 27235449 Free PMC article. * The genetics of heart attack. Topol EJ. Topol EJ. Heart. 2006 Jun;92(6):855-61. doi: 10.1136/hrt.2005.060202. Heart. 2006. PMID: 16698843 Free PMC article. Review. No abstract available. * Lack of MEF2A Delta7aa mutation in Irish families with early onset ischaemic heart disease, a family based study. Horan PG, Allen AR, Hughes AE, Patterson CC, Spence M, McGlinchey PG, Belton C, Jardine TC, McKeown PP. Horan PG, et al. BMC Med Genet. 2006 Jul 27;7:65. doi: 10.1186/1471-2350-7-65. BMC Med Genet. 2006. PMID: 16872533 Free PMC article. * A clinical approach to inherited premature coronary artery disease. Stitziel NO, MacRae CA. Stitziel NO, et al. Circ Cardiovasc Genet. 2014 Aug;7(4):558-64. doi: 10.1161/CIRCGENETICS.113.000152. Circ Cardiovasc Genet. 2014. PMID: 25140063 Free PMC article. No abstract available. * Pathogenesis of coronary artery disease: focus on genetic risk factors and identification of genetic variants. Sayols-Baixeras S, Lluís-Ganella C, Lucas G, Elosua R. Sayols-Baixeras S, et al. Appl Clin Genet. 2014 Jan 16;7:15-32. doi: 10.2147/TACG.S35301. eCollection 2014. Appl Clin Genet. 2014. PMID: 24520200 Free PMC article. Review.
## References
1. 1. Murray CJ, Lopez AD. Lancet. 1997;349:1498. - PubMed 2. 1. Lusis AJ. Nature. 2000;407:233. - PMC - PubMed 3. 1. Nora JJ, et al. Circulation. 1980;61:503. - PubMed 4. 1. Schildkraut JM, Myers RH, Cupples LA, Kiely DK, Kannel WB. Am J Cardiol. 1989;64:555. - PubMed 5. 1. Wang Q, Pyeritz RE. In: Textbook of Cardiovascular Medicine. EJ Topol., editor. Vol. 3. Lippincott, Williams & Wilkins; New York: 2000. pp. 1–13.
## Publication types
## MeSH terms
## Substances
## Grants and funding
* R01 HL065630/HL/NHLBI NIH HHS/United States * R01 HL066251/HL/NHLBI NIH HHS/United States * R01 HL65630/HL/NHLBI NIH HHS/United States * R01 HL66251/HL/NHLBI NIH HHS/United States
## LinkOut - more resources
* ### Full Text Sources * Atypon * Europe PubMed Central * Nature Publishing Group * Ovid Technologies, Inc. * PubMed Central * ### Other Literature Sources * H1 Connect - Access expert opinions and insights on biomedical research. * The Lens - Patent Citations Database * ### Medical * MedlinePlus Health Information * ### Miscellaneous * NCI CPTAC Assay Portal