Clone-based systematic haplotyping (CSH): a procedure for physical haplotyping of whole genomes - PubMed (original) (raw)
Clone-based systematic haplotyping (CSH): a procedure for physical haplotyping of whole genomes
Carola Burgtorf et al. Genome Res. 2003 Dec.
Erratum in
- Genome Res. 2004 Feb;14(4):327
Abstract
We present a novel methodology to determine the phase of single-nucleotide polymorphisms (SNPs) on a chromosome, which we term clone-based systematic haplotyping (CSH). The CSH procedure is based on separating the allelic chromosomes of a diploid genome by fosmid/cosmid cloning, and subsequent SNP typing of 96 clone pools, each representing approximately 10% of the genome. The pools are screened by PCR for the sequence of interest, followed by SNP typing on the PCR products using the GOOD assay. We demonstrate that by CSH, the haplotype of SNPs separated by more than 50 kilobases can definitely be assigned. We propose this method as being suitable for constructing maps of ancestral haplotypes, analysis of complex diseases, and for diagnosis of rare defects in which the molecular haplotype is crucial. In addition, by amplifying the initial DNA by many orders of magnitude, the original DNA resource is effectively immortalized, enabling the haplotyping of hundreds of thousands of SNPs per individual.
Figures
Figure 2
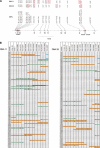
(A) Schematic drawing of the OPRM1 genomic region. SNPs are indicated and tagged with the published names (NCBI dbSNP-database; Hoehe et al. 2000). The genotypes of DNA11 and DNA28 are shown in the top lines. (B) Results for the haplotyping of DNA11 and DNA28. The different haplotypes are represented in red and green, respectively. A“+” indicates positive a PCR of SNPs that were previously found to be homozygous for DNA11 and DNA28, respectively. The letters A, C, G, and T denote the result of the haplotyping by the GOOD assay of the pools analyzed. Blue bars: pools where only homozygous SNPs are detected. Brown indicates the presence of two clones of different haplotypes in one pool.
Figure 1
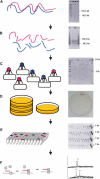
(Left) A schematic representation of the CSH method; (right) a corresponding example of “real data.” (A) Intact high-molecular-weight DNA is prepared, and the quality is checked by PFG electrophoresis. (B) DNA is sheared into pieces of ∼40–100 kb, as can be seen by PFG. (C) After end-repair, a cosmid/fosmid library is constructed by ligation of the fragments to a suitable vector, in vitro packaging, and transfection of bacteria. The quality of the library is checked by restriction analysis of a few randomly chosen clones. (D) The titer of the library is determined, and ∼10,000 clones are plated onto one plate. (E) The region of interest is amplified by PCR, and the positive pools are selected for subsequent analysis by the GOOD assay; as an example PCR3 of DNA11 is shown. (F) The genotypes of the SNPs are analyzed using the GOOD assay (Fig. 3).
Figure 3
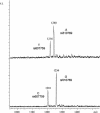
A duplex haplotype analysis of two SNPs lying 10 kb apart. For SNP rs510769, primer TATGGCATTTCACATTCACATGptTA was used; for SNP rs607759 of the OPRM1 gene primer, AATTGAATGGCTCTAGGptAC was used. Respective products for rs510769 were Gpt-TApt(G/A) with masses 1256 Da and 1240 Da, and respective products for rs607759 were GptAC(T/C) with masses 1216 and 1201. Top trace: The mass spectrometry analysis of SNPs rs510769 and rs607759 of the OPRM1 gene in a specific fosmid pool. As can be deduced from the masses, the A allele of SNP rs510769 and the T allele of SNP rs607759 were in phase. Bottom trace: The analysis of the complementary fosmid pool, where the G allele of SNP rs510769 and the C allele of SNP rs607759 were in phase.
Figure 4
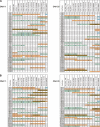
The effect of larger pools was simulated `electronically'. (A) Two pools were combined; (B) three pools.
Similar articles
- Targeted, haplotype-resolved resequencing of long segments of the human genome.
Raymond CK, Subramanian S, Paddock M, Qiu R, Deodato C, Palmieri A, Chang J, Radke T, Haugen E, Kas A, Waring D, Bovee D, Stacy R, Kaul R, Olson MV. Raymond CK, et al. Genomics. 2005 Dec;86(6):759-66. doi: 10.1016/j.ygeno.2005.08.013. Epub 2005 Oct 24. Genomics. 2005. PMID: 16249066 - [Analysis and application of SNP and haplotype in the human genome].
Li J, Pan YC, Li YX, Shi TL. Li J, et al. Yi Chuan Xue Bao. 2005 Aug;32(8):879-89. Yi Chuan Xue Bao. 2005. PMID: 16231744 Review. Chinese. - Effects of single SNPs, haplotypes, and whole-genome LD maps on accuracy of association mapping.
Maniatis N, Collins A, Morton NE. Maniatis N, et al. Genet Epidemiol. 2007 Apr;31(3):179-88. doi: 10.1002/gepi.20199. Genet Epidemiol. 2007. PMID: 17285621 - A Fosmid Pool-Based Next Generation Sequencing Approach to Haplotype-Resolve Whole Genomes.
Suk EK, Schulz S, Mentrup B, Huebsch T, Duitama J, Hoehe MR. Suk EK, et al. Methods Mol Biol. 2017;1551:223-269. doi: 10.1007/978-1-4939-6750-6_13. Methods Mol Biol. 2017. PMID: 28138850 - Tag SNP selection for association studies.
Stram DO. Stram DO. Genet Epidemiol. 2004 Dec;27(4):365-74. doi: 10.1002/gepi.20028. Genet Epidemiol. 2004. PMID: 15372618 Review.
Cited by
- Experimental generation of SNP haplotype signatures in patients with sickle cell anaemia.
Menzel S, Qin J, Vasavda N, Thein SL, Ramakrishnan R. Menzel S, et al. PLoS One. 2010 Sep 24;5(9):e13004. doi: 10.1371/journal.pone.0013004. PLoS One. 2010. PMID: 20886046 Free PMC article. - Long-range multilocus haplotype phasing of the MHC.
Guo Z, Hood L, Malkki M, Petersdorf EW. Guo Z, et al. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6964-9. doi: 10.1073/pnas.0602286103. Epub 2006 Apr 21. Proc Natl Acad Sci U S A. 2006. PMID: 16632595 Free PMC article. - Molecular haplotyping by linking emulsion PCR: analysis of paraoxonase 1 haplotypes and phenotypes.
Wetmur JG, Kumar M, Zhang L, Palomeque C, Wallenstein S, Chen J. Wetmur JG, et al. Nucleic Acids Res. 2005 May 10;33(8):2615-9. doi: 10.1093/nar/gki556. Print 2005. Nucleic Acids Res. 2005. PMID: 15886392 Free PMC article. - Recent Advances in Experimental Whole Genome Haplotyping Methods.
Huang M, Tu J, Lu Z. Huang M, et al. Int J Mol Sci. 2017 Sep 11;18(9):1944. doi: 10.3390/ijms18091944. Int J Mol Sci. 2017. PMID: 28891974 Free PMC article. Review. - Whole-genome molecular haplotyping of single cells.
Fan HC, Wang J, Potanina A, Quake SR. Fan HC, et al. Nat Biotechnol. 2011 Jan;29(1):51-7. doi: 10.1038/nbt.1739. Epub 2010 Dec 19. Nat Biotechnol. 2011. PMID: 21170043 Free PMC article.
References
- Boldt, A.B. and Petzl-Erler, M.L. 2002. A new strategy for mannose-binding lectin gene haplotyping. Hum. Mutat. 19: 296-306. - PubMed
- Daly, M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J., and Lander, E.S. 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229-232. - PubMed
- Davidson, S. 2000. Research suggests importance of haplotypes over SNPs. Nat. Biotechnol. 18: 1134-1135. - PubMed
- Douglas, J.A., Boehnke, M., Gillanders, E., Trent, J.M., and Gruber, S.B. 2001. Experimentally-derived haplotypes substantially increase the efficiency of linkage disequilibrium studies. Nat. Genet. 28: 361-364. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources