Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S - PubMed (original) (raw)
Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S
David J Katzmann et al. Mol Biol Cell. 2004 Feb.
Abstract
The multivesicular body (MVB) sorting pathway provides a mechanism for delivering transmembrane proteins into the lumen of the lysosome/vacuole. Recent studies demonstrated that ubiquitin modification acts in cis as a signal for the sorting of cargoes into this pathway. Here, we present results from a genetic selection designed to identify mutants that missort MVB cargoes. This selection identified a point mutation in ubiquitin ligase Rsp5 (Rsp5-326). At the permissive temperature, this mutant is specifically defective for ubiquitination and sorting of the ubiquitin-dependent MVB cargo precursor carboxypeptidase S (pCPS), but not ligand-induced ubiquitination of Ste2. A previous study implicated Tul1 as the ubiquitin ligase responsible for MVB sorting of pCPS. However, we detected no defect in either the sorting or ubiquitination of pCPS in tul1 mutants. We had previously shown that Fab1 phosphatidylinositol 3-phosphate 5-kinase is also required for MVB sorting of pCPS, but not Ste2. However, our analyses reveal that fab1 mutants do not exhibit a defect in ubiquitination of pCPS. Thus, both Rsp5 and Fab1 play distinct and essential roles in the targeting of biosynthetic MVB cargoes. However, whereas Rsp5 seems to be responsible for cargo ubiquitination, the precise role for Fab1 remains to be elucidated.
Figures
Figure 1.
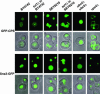
mvb326 cells fail to exhibit a class E compartment but specifically mislocalize the MVB cargo GFP-CPS. (A) Vacuole morphology of mvb326 and MBY3 (_vps4_Δ) cells was visualized by either staining with FM 4-64 (left) or by using Nomarski optics (right). Cells were grown to midlog phase at 26°C, stained with FM 4-64 for 20 min, and chased with YPD for 1 h. (B) Localization of GFP-CPS, Ub-GFP-CPS, and _Sna_III-GFP in SEY6210 (wild-type) and mvb326 at 26°C.
Figure 2.
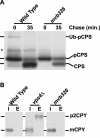
mvb326 cells exhibit a defect in ubiquitination of pCPS. (A) CPS was immunoprecipitated from either CBY16 (_pep12_Δ) or SSY25 (_mvb326 pep12_Δ) cells, grown at 26°C, followed by Western blot analysis by using anti-ubiquitin antibody. (B) SEY6210 (wild-type), GOY19 (_cps1_Δ), and mvb326 cells were labeled with Trans 35S-label for 10 min at 26°C. Labeling was terminated by addition of unlabeled methionine and cysteine, and CPS was immunoprecipitated from the extracts. Before SDS-PAGE analysis, extracts were deglycosylated by treatment with endoglycosidase H, which resulted in only a single band for Ub-pCPS. An asterisk highlights a nonspecific band.
Figure 3.
mvb326 cells exhibit a specific defect in pCPS processing. (A) SEY6210 (wild-type) and mvb326 cells were labeled with Trans 35S-label for 10 min at 26°C and chased with unlabeled methionine and cysteine for 35 min. Extracts were processed as in Figure 2. An asterisk highlights a nonspecific band as described in Figure 2. (B) SEY6210 (wild-type), MBY3 (_vps4_Δ), and mvb326 cells were grown at 26°C and converted to spheroplasts. The spheroplasts were labeled with Trans 35S-label for 10 min at 26°C and then chased for 30 min by using unlabeled methionine and cysteine. Spheroplasts were then harvested and separated into intracellular (I) or extracellular (E) fractions, and CPY was immunoprecipitated from each fraction.
Figure 4.
Entry into the MVB pathway is not affected in _tul1_Δ or _ubc4_Δ ubc5Δ cells. Localization of GFP-CPS (top) and _Sna_III-GFP (bottom) in BY4742 (wild-type), 14833 (_tul1_Δ in BY4742), SEY6210 (wild-type), SSY11 (_tul1_Δ in SEY6210), LH21 (_ubc4_Δ ubc5Δ), and MBY3 (_vps4_Δ) cells.
Figure 5.
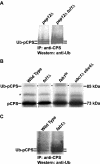
pCPS is ubiquitinated in _tul1_Δ, _ubc1_Δ ubc4Δ, and fab1 cells. (A) CPS was immunoprecipitated from either CBY16 (_pep12_Δ) or SSY16 (_tul1_Δ pep12Δ) cells, grown at 26°C, followed by Western blot analysis by using anti-ubiquitin antibody. (B) SEY6210 (wild-type), SSY11 (_tul1_Δ), and LH183 (_ubc1_Δ ubc4Δ) cells were labeled with Trans 35S-label for 10 min at 26°C and chased with unlabeled methionine and cysteine. Extracts ere prepared and immunoprecipitated for CPS as described in Figure 2. EMY119 (fab1ts) cells were treated similarly except for a brief shift to 38°C (15 min) before labeling. An asterisk highlights a nonspecific band as described in Figure 2. (C) CPS was immunoprecipitated from either SEY6210 (wild-type) or _fab1_Δ1 (_fab1_Δ) cells, grown at 26°C, followed by Western blot analysis by using anti-ubiquitin antibody.
Figure 6.
Specific alleles of RSP5 differentially affect pCPS entry into the MVB pathway and ubiquitination of Ste2. (A) Localization of GFP-CPS in mvb326 cells, harboring a plasmid encoding HA-Rsp5 (top), or _rsp5_Δ cells harboring a plasmid encoding Rsp5-326 (bottom), at 26°C. (B) SEY6211 (wild-type) and SSY22 (mvb326) cells expressing Ste2-HA were treated with alpha factor for 8 min at the indicated temperatures and lysates were prepared and probed with anti-HA antibody. Unmodified Ste2, phosphorylated Ste2, and ubiquitinated Ste2 are indicated by brackets. (C) CPS was immunoprecipitated from either CBY16 (_pep12_Δ) or DKY83 (_smm1 pep12_Δ) cells, grown at 26°C, followed by Western blot analysis by using anti-ubiquitin antibody. (D) LH291 (wild-type), LH23 (rsp5-1) MYY290 (wild-type) and MYY833 (mdp1-13) cells were incubated at 37°C for 10 min, before labeling with Trans 35S-label for 10 min at 37°C. Cells were treated as described in Figure 2. An asterisk marks a nonspecific band as described in Figure 2.
Figure 7.
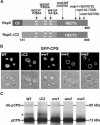
The C2 and WW domains of Rsp5 also play a role in the ubiquitination and sorting of pCPS. (A) Schematic of Rsp5 domain structure with mutations mapped to each domain. (B) Localization of GFPCPS in SEY6210 (wild-type), TCY124 (ΔC2), TCY84 (ww1), TCY86 (ww2), and TCY81 (ww3) mutants. (C) SEY6210 (wild-type), TCY124 (ΔC2), TCY84 (ww1), TCY86 (ww2), and TCY81 (ww3) cells were labeled with Trans 35S-label for 10 min at 26°C and chased with unlabeled methionine and cysteine. Extracts were processed as in Figure 2. An asterisk highlights a nonspecific band as described in Figure 2.
Figure 8.
Localization of GFP-Rsp5. SEY6210 cells expressing either GFP-Rsp5 and Sec7-DsRed (top) or GFP-Rsp5 and DsRed-FYVE (bottom) were grown at 26°C, and the GFP and DsRed labeled proteins were visualized by fluorescence microscopy.
Similar articles
- Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5.
Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Oestreich AJ, et al. Mol Biol Cell. 2007 Feb;18(2):707-20. doi: 10.1091/mbc.e06-08-0680. Epub 2006 Dec 20. Mol Biol Cell. 2007. PMID: 17182849 Free PMC article. - Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation.
Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P. Stawiecka-Mirota M, et al. Traffic. 2007 Sep;8(9):1280-96. doi: 10.1111/j.1600-0854.2007.00610.x. Epub 2007 Jul 23. Traffic. 2007. PMID: 17645729 Free PMC article. - Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies.
Ren J, Kee Y, Huibregtse JM, Piper RC. Ren J, et al. Mol Biol Cell. 2007 Jan;18(1):324-35. doi: 10.1091/mbc.e06-06-0557. Epub 2006 Nov 1. Mol Biol Cell. 2007. PMID: 17079730 Free PMC article. - Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking.
Belgareh-Touzé N, Léon S, Erpapazoglou Z, Stawiecka-Mirota M, Urban-Grimal D, Haguenauer-Tsapis R. Belgareh-Touzé N, et al. Biochem Soc Trans. 2008 Oct;36(Pt 5):791-6. doi: 10.1042/BST0360791. Biochem Soc Trans. 2008. PMID: 18793138 Review. - The ubiquitin code of yeast permease trafficking.
Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. Lauwers E, et al. Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
Cited by
- Identification of putative effectors of the Type IV secretion system from the Wolbachia endosymbiont of Brugia malayi.
Carpinone EM, Li Z, Mills MK, Foltz C, Brannon ER, Carlow CKS, Starai VJ. Carpinone EM, et al. PLoS One. 2018 Sep 27;13(9):e0204736. doi: 10.1371/journal.pone.0204736. eCollection 2018. PLoS One. 2018. PMID: 30261054 Free PMC article. - A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting.
Erpapazoglou Z, Dhaoui M, Pantazopoulou M, Giordano F, Mari M, Léon S, Raposo G, Reggiori F, Haguenauer-Tsapis R. Erpapazoglou Z, et al. Mol Biol Cell. 2012 Jun;23(11):2170-83. doi: 10.1091/mbc.E11-10-0891. Epub 2012 Apr 4. Mol Biol Cell. 2012. PMID: 22493318 Free PMC article. - Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch.
Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. Jadwin JA, et al. J Virol. 2010 Jan;84(2):704-15. doi: 10.1128/JVI.01319-09. Epub 2009 Oct 28. J Virol. 2010. PMID: 19864377 Free PMC article. - Membrane protein targeting to the MVB/lysosome.
Davies BA, Lee JR, Oestreich AJ, Katzmann DJ. Davies BA, et al. Chem Rev. 2009 Apr;109(4):1575-86. doi: 10.1021/cr800473s. Chem Rev. 2009. PMID: 19243135 Free PMC article. Review. No abstract available. - COP9-associated CSN5 regulates exosomal protein deubiquitination and sorting.
Liu Y, Shah SV, Xiang X, Wang J, Deng ZB, Liu C, Zhang L, Wu J, Edmonds T, Jambor C, Kappes JC, Zhang HG. Liu Y, et al. Am J Pathol. 2009 Apr;174(4):1415-25. doi: 10.2353/ajpath.2009.080861. Epub 2009 Feb 26. Am J Pathol. 2009. PMID: 19246649 Free PMC article.
References
- Babst, M., Katzmann, D.J., Estepa-Sabal, E.J., Meerloo, T., and Emr, S.D. (2002a). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3, 271-282. - PubMed
- Babst, M., Katzmann, D.J., Snyder, W.B., Wendland, B., and Emr, S.D. (2002b). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283-289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases