MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation - PubMed (original) (raw)
MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation
Yi Wang et al. Proc Natl Acad Sci U S A. 2003.
Abstract
The mismatch repair proteins function upstream in the DNA damage signaling pathways induced by the DNA methylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG). We report that MSH2 (MutS homolog 2) protein interacts with the ATR (ATM- and Rad3-related) kinase to form a signaling module and regulate the phosphorylation of Chk1 and SMC1 (structure maintenance of chromosome 1). We found that phosphorylation of Chk1 by ATR also requires checkpoint proteins Rad17 and replication protein A. In contrast, phosphorylation of SMC1 by ATR is independent of Rad17 and replication protein A, suggesting that the signaling pathway leading to SMC1 phosphorylation is distinct from that mediated by the checkpoint proteins. In addition, both MSH2 and Rad17 are required for the activation of the S-phase checkpoint to suppress DNA synthesis in response to MNNG, and phosphorylation of SMC1 is required for cellular survival. These data support a model in which MSH2 and ATR function upstream to regulate two branches of the response pathway to DNA damage caused by MNNG.
Figures
Fig. 1.
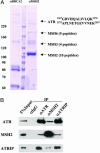
MSH2 associates with ATR in HeLa NE. (A) The MSH2 complex was isolated by anti-MSH2 IP and separated on a SDS/PAGE. A parallel IP using a BRCA2 antibody serves as a negative control. Protein bands stained by Coomassie blue were analyzed by mass spectrometry. The sequences of the two ATR peptides identified are shown. (B) Coimmunoprecipitations of MSH2, ATR, and ATRIP detected by Western blotting. IPs were carried out in HeLa NE by using indicated antibodies. Five percent of the total protein used in the IP was loaded in the input lane.
Fig. 2.
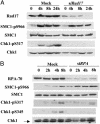
The MSH2/ATR complex is required for MNNG-induced phosphorylation of SMC1 and Chk1. (A) In vitro kinase assays of SMC1 by ATR. Transiently expressed Flag-tagged ATR (wild type or kinase dead) in 293T cells was used to phosphorylate a GST-SMC1 fragment (amino acids 890-1233). The phosphorylation product was immunoblotted with SMC1 S966 phosphospecific antibody. Equal amounts of GST-SMC1 proteins used in the kinase assays were monitored by Coomassie blue staining. (B and C) Dependence of SMC1 S966 and Chk1 S317 phosphorylations on MSH2 (B) and ATR (C) in response to MNNG. HeLa cells transfected with mock (siGFP), si_ATR_, or si_MSH2_ were treated with 10 μM MNNG for 1 h and harvested at indicated times. Efficiency of RNA interference was monitored by Western blotting to each protein. (D) ATR, not ATM, is the major kinase involved in early response to MNNG-induced damage. Mock, siRNA to _ATM_-or _ATR_-transfected HeLa cells was treated with 10 μM MNNG for 1 h and harvested after 4 h. Cell lysates were analyzed with indicated antibodies. Chk1 blot also serves as a loading control. (E) Independence of SMC1 phosphorylation on ATR and MSH2 in response to γ irradiation. HeLa cells transfected with indicated siRNA were treated with 10 Gy of IR and harvested after 1 h.
Fig. 3.
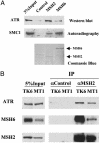
Interaction of MSH2 or MSH6 with ATR and SMC1. (A) In vitro interactions of MSH2 or MSH6 with ATR and SMC1, respectively. Affinity-purified human MSH2 and MSH6 from sf9 cells were incubated with purified Flag-ATR or 35S-labeled, in vitro translated SMC1. An unrelated antibody incubated with sf9 cell lysate was used as a control. Coomassie blue staining of the membrane after Western blotting shows that human MSH2 and MSH6 used in the binding reactions do not copurify with the insect MSH6 or MSH2. (B) MSH6-independent association of MSH2 and ATR. IPs were carried out in NE prepared from lymphoblastoid cell line TK6, which expresses wild-type MSH6, and MT1, which expresses a low level of a mutant form of MSH6.
Fig. 4.
Rad17 and RPA are not required for the phosphorylation of SMC1. Shown is MNNG-induced phosphorylation of SMC1 and Chk1 in the absence of Rad17 (A) or RPA (B). HeLa cells transfected with indicated siRNAs were treated with 10 μM MNNG for 1 h and harvested at indicated times. Lysates were analyzed with indicated antibodies.
Fig. 5.
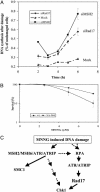
(A) Both MSH2 and Rad17 are required for activation of MNNG-induced S-phase checkpoint. HeLa cells transfected with mock, MSH2, or Rad17 siRNA were treated with 10 μM MNNG for 1 h. DNA synthesis measured by [3H]thymidine incorporation was evaluated at time indicated after the treatment and normalized to that in cycling cells. The measurements were performed in triplicate. The error bar represents the standard deviation. (B) SMC1 phosphorylation is required for cellular survival after MNNG treatment. HeLa cells expressing either GFP-wt-SMC1 or GFP-SA966-SMC1 were treated with MNNG of indicated concentrations for 1 h and recovered for 1 week before assaying for colony formation. The assay was performed in triplicate. The error bar represents the standard deviation. (C) A schematic model of MNNG-induced DNA damage response. Upon exposure to MNNG, binding of the MSH2/MSH6 heterodimer to O6-methyl-G·C activates the checkpoint kinase ATR, leading to the phosphorylation of SMC1 and Chk1. The RPA is also required to recruit the ATR/ATRIP complex to the damaged sites, possibly through binding to MMR intermediates, resulting in Rad17-dependent phosphorylation of Chk1. However, SMC1 can be phosphorylated independent of RPA and Rad17, suggesting the existence of a response pathway through direct interaction between ATR/MSH2 and MSH6/SMC1.
Similar articles
- The alkylating carcinogen N-methyl-N'-nitro-N-nitrosoguanidine activates the plasminogen activator inhibitor-1 gene through sequential phosphorylation of p53 by ATM and ATR kinases.
Vidal B, Parra M, Jardí M, Saito S, Appella E, Muñoz-Cánoves P. Vidal B, et al. Thromb Haemost. 2005 Mar;93(3):584-91. doi: 10.1160/TH04-10-0644. Thromb Haemost. 2005. PMID: 15735814 - Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase.
Stojic L, Mojas N, Cejka P, Di Pietro M, Ferrari S, Marra G, Jiricny J. Stojic L, et al. Genes Dev. 2004 Jun 1;18(11):1331-44. doi: 10.1101/gad.294404. Genes Dev. 2004. PMID: 15175264 Free PMC article. - Methylator-induced, mismatch repair-dependent G2 arrest is activated through Chk1 and Chk2.
Adamson AW, Beardsley DI, Kim WJ, Gao Y, Baskaran R, Brown KD. Adamson AW, et al. Mol Biol Cell. 2005 Mar;16(3):1513-26. doi: 10.1091/mbc.e04-02-0089. Epub 2005 Jan 12. Mol Biol Cell. 2005. PMID: 15647386 Free PMC article. - The ATM-dependent DNA damage signaling pathway.
Kitagawa R, Kastan MB. Kitagawa R, et al. Cold Spring Harb Symp Quant Biol. 2005;70:99-109. doi: 10.1101/sqb.2005.70.002. Cold Spring Harb Symp Quant Biol. 2005. PMID: 16869743 Review. - ATR signalling: more than meeting at the fork.
Nam EA, Cortez D. Nam EA, et al. Biochem J. 2011 Jun 15;436(3):527-36. doi: 10.1042/BJ20102162. Biochem J. 2011. PMID: 21615334 Free PMC article. Review.
Cited by
- Exonuclease 1 (Exo1) is required for activating response to S(N)1 DNA methylating agents.
Izumchenko E, Saydi J, Brown KD. Izumchenko E, et al. DNA Repair (Amst). 2012 Dec 1;11(12):951-64. doi: 10.1016/j.dnarep.2012.09.004. Epub 2012 Oct 11. DNA Repair (Amst). 2012. PMID: 23062884 Free PMC article. - Protein phosphatase 5 is required for ATR-mediated checkpoint activation.
Zhang J, Bao S, Furumai R, Kucera KS, Ali A, Dean NM, Wang XF. Zhang J, et al. Mol Cell Biol. 2005 Nov;25(22):9910-9. doi: 10.1128/MCB.25.22.9910-9919.2005. Mol Cell Biol. 2005. PMID: 16260606 Free PMC article. - TLR9 engagement on CD4 T lymphocytes represses gamma-radiation-induced apoptosis through activation of checkpoint kinase response elements.
Zheng L, Asprodites N, Keene AH, Rodriguez P, Brown KD, Davila E. Zheng L, et al. Blood. 2008 Mar 1;111(5):2704-13. doi: 10.1182/blood-2007-07-104141. Epub 2007 Dec 17. Blood. 2008. PMID: 18086870 Free PMC article. - N-nitroso-N-ethylurea activates DNA damage surveillance pathways and induces transformation in mammalian cells.
Bodakuntla S, Libi AV, Sural S, Trivedi P, Lahiri M. Bodakuntla S, et al. BMC Cancer. 2014 Apr 24;14:287. doi: 10.1186/1471-2407-14-287. BMC Cancer. 2014. PMID: 24758542 Free PMC article. - ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts.
Yoshioka K, Yoshioka Y, Hsieh P. Yoshioka K, et al. Mol Cell. 2006 May 19;22(4):501-10. doi: 10.1016/j.molcel.2006.04.023. Mol Cell. 2006. PMID: 16713580 Free PMC article.
References
- Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397–408. - PubMed
- Lydall, D. & Weinert, T. (1995) Science 270, 1488–1491. - PubMed
- Zou, L. & Elledge, S. J. (2003) Science 300, 1542–1548. - PubMed
- Friedberg, E. C., Walker, G. C. & Sinclair, D. A. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC.).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous