Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides - PubMed (original) (raw)
Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides
Hamilton O Smith et al. Proc Natl Acad Sci U S A. 2003.
Abstract
We have improved upon the methodology and dramatically shortened the time required for accurate assembly of 5- to 6-kb segments of DNA from synthetic oligonucleotides. As a test of this methodology, we have established conditions for the rapid (14-day) assembly of the complete infectious genome of bacteriophage X174 (5386 bp) from a single pool of chemically synthesized oligonucleotides. The procedure involves three key steps: (i). gel purification of pooled oligonucleotides to reduce contamination with molecules of incorrect chain length, (ii). ligation of the oligonucleotides under stringent annealing conditions (55 degrees C) to select against annealing of molecules with incorrect sequences, and (iii). assembly of ligation products into full-length genomes by polymerase cycling assembly, a nonexponential reaction in which each terminal oligonucleotide can be extended only once to produce a full-length molecule. We observed a discrete band of full-length assemblies upon gel analysis of the polymerase cycling assembly product, without any PCR amplification. PCR amplification was then used to obtain larger amounts of pure full-length genomes for circularization and infectivity measurements. The synthetic DNA had a lower infectivity than natural DNA, indicating approximately one lethal error per 500 bp. However, fully infectious X174 virions were recovered after electroporation into Escherichia coli. Sequence analysis of several infectious isolates verified the accuracy of these synthetic genomes. One such isolate had exactly the intended sequence. We propose to assemble larger genomes by joining separately assembled 5- to 6-kb segments; approximately 60 such segments would be required for a minimal cellular genome.
Figures
Fig. 1.
Schematic diagram of the steps in the global synthesis of infectious φX174 bacteriophage from synthetic oligonucleotides.
Fig. 2.
(A) Computer simulation of the ligation reaction as a function of variation in the concentrations of the input oligonucleotides. Computer simulation parameters are 130 top oligonucleotides (with bottom oligonucleotides saturating) and 2,000 molecules of each top oligonucleotide. Percent variation of oligonucleotide concentrations is determined by the randomly chosen number of molecules of each oligonucleotide. For example, at 10% variation in oligonucleotide concentrations, the number of each oligonucleotide is randomly chosen between 1,800 and 2,000. At each iteration of the program, a random pair of assemblies is selected and the pairs are joined together to form a larger assembly if the end coordinate of one assembly is one less than the beginning of the other assembly. The process is iterated until the number of assemblies no longer changes during 1 million pairings. -▪-▪-, average percent length of assemblies; -•-•-, percent of full-length assemblies. (B) The final products of a theoretical PCA reaction that starts with oligonucleotides of uniform size. Only the two terminal oligonucleotides can be extended to full length because polymerization is only in the 5′ to 3′ direction. An assembly is considered “active” if it can be extended by overlapping with another assembly followed by fill-in synthesis. An assembly is “inactive” if it cannot be extended further. Mass increase factor = final mass/beginning mass = sn (n + 2)/2_sn_ = (n + 2)/2 ≈ n/2. Fraction of final mass full length = [2 (sn + s/2)]/[sn (n + 2)] = [2 (n + 1/2)]/[n (n + 2)]≈ 2/n. s = nucleotide length of oligonucleotide; n = number of oligonucleotides on one strand.
Fig. 3.
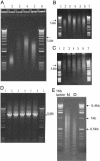
PCA of full-length synφX molecules. The first stage of PCA (50-μl reaction volume) was carried out for 35 cycles with 0.2, 0.5, or 1 μlofthe Taq ligation product. PCA products were analyzed on a 0.8% E-gel. (A) Lanes 1 and 6, 1-kb ladder; lane 2, 0.5 μlof Taq ligation product; lane 3, 2 μl of the 0.2-μl PCA; lane 4, 2 μl of the 0.5-μl PCA; lane 5, 2 μl of the 1-μl PCA. The second stage of PCA was for an additional 35 cycles in five new 50-μl reactions. For reaction 1, the 0.2-μl first-stage reaction was continued without change for another 35 cycles with the addition of 0.5 μl of fresh HF polymerase mixture. For reactions 2 and 3, 10 and 20 μl of the 0.5-μl first-stage PCA product was used. For reactions 4 and 5, 5 and 10 μl of the 1-μl first-stage PCA product was used. Analysis was on 0.8% E-gels. (B) Formamide-denatured DNA. (C) Native DNA. (B and C) Lanes 1 and 7, 1-kb ladder; lane 2, 2 μl of reaction 1; lanes 3 and 4, 2 μl of reactions 2 and 3; lanes 5 and 6, 2 μl of reactions 4 and 5. (D) PCR amplification of the products of the second set of PCA products as shown in B and C.(E) Taq ligase assembly of 259 oligonucleotides. A 0.5-μl sample of the ligation products was analyzed on a 2% E-gel (Invitrogen) in duplex form (lane N). One microliter of the ligation products was mixed with 20 μl of formamide, heated to 95°C for 2 min, and then analyzed (lane D). Denatured standards run approximately the same as native standards, based on other experiments (data not shown).
Fig. 4.
Plaques of synφX-A. There appear to be several plaque morphologies: small plaques with sharp borders, medium-sized plaques, and large plaques with fuzzy borders.
Fig. 5.
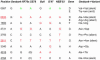
Sequence comparisons of natural φX and synφX genomes. Differences from the Sanger sequence (13) are indicated. A4, A8, B1, and B3 are the synφX described in the text. NEB, φX RF I DNA supplied by NEB (catalog no. N3021S); RF70s, DNA prepared in the late 1970s and stored since then by C.A.H.
Fig. 6.
Sequence differences between the Sanger sequence and more recent sequencing of natural φX DNAs. RF70s, a preparation of φX double-stranded RF from the late 1970s; SS78, a preparation of φX virion single-stranded DNA from 1978; Bull, the sequence of wild-type φX used by Bull et al. (15); G'97, φX RF DNA from 1997; NEB'03, φX RF DNA from NEB in use at the Institute for Biological Energy Alternatives during the φX genome synthesis.
Similar articles
- PCR-based accurate synthesis of long DNA sequences.
Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ, Cheng ZM, Li Y. Xiong AS, et al. Nat Protoc. 2006;1(2):791-7. doi: 10.1038/nprot.2006.103. Nat Protoc. 2006. PMID: 17406309 - Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome.
Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA 3rd, Smith HO. Gibson DG, et al. Science. 2008 Feb 29;319(5867):1215-20. doi: 10.1126/science.1151721. Epub 2008 Jan 24. Science. 2008. PMID: 18218864 - A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast.
Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. Jaschke PR, et al. Virology. 2012 Dec 20;434(2):278-84. doi: 10.1016/j.virol.2012.09.020. Epub 2012 Oct 15. Virology. 2012. PMID: 23079106 - Another side of genomics: synthetic biology as a means for the exploitation of whole-genome sequence information.
Stähler P, Beier M, Gao X, Hoheisel JD. Stähler P, et al. J Biotechnol. 2006 Jun 25;124(1):206-12. doi: 10.1016/j.jbiotec.2005.12.011. Epub 2006 Jan 24. J Biotechnol. 2006. PMID: 16436303 Review. - [Synthesis of artifical genome as the basis of synthetic biology].
Rad'ko SP, Il'ina AP, Bodoev NV, Archakov AI. Rad'ko SP, et al. Biomed Khim. 2007 May-Jun;53(3):237-48. Biomed Khim. 2007. PMID: 17722574 Review. Russian.
Cited by
- The design and engineering of synthetic genomes.
James JS, Dai J, Chew WL, Cai Y. James JS, et al. Nat Rev Genet. 2024 Nov 6. doi: 10.1038/s41576-024-00786-y. Online ahead of print. Nat Rev Genet. 2024. PMID: 39506144 Review. - Synthetic moss.
[No authors listed] [No authors listed] Nat Plants. 2024 Jun;10(6):831-832. doi: 10.1038/s41477-024-01733-9. Nat Plants. 2024. PMID: 38906995 No abstract available. - Designing a synthetic moss genome using GenoDesigner.
Yu W, Zhang S, Zhao S, Chen LG, Cao J, Ye H, Yan J, Zhao Q, Mo B, Wang Y, Jiao Y, Ma Y, Huang X, Qian W, Dai J. Yu W, et al. Nat Plants. 2024 Jun;10(6):848-856. doi: 10.1038/s41477-024-01693-0. Epub 2024 Jun 3. Nat Plants. 2024. PMID: 38831044 Review. - Synthetic Genomics: Repurposing Biological Systems for Applications in Engineering Biology.
Fu X, Shen Y. Fu X, et al. ACS Synth Biol. 2024 May 17;13(5):1394-1399. doi: 10.1021/acssynbio.4c00006. ACS Synth Biol. 2024. PMID: 38757697 Free PMC article. Review. - POSoligo software for in vitro gene synthesis.
Tong Y, Sun J, Chen Y, Yi C, Wang H, Li C, Dai N, Yang G. Tong Y, et al. Sci Rep. 2024 May 15;14(1):11117. doi: 10.1038/s41598-024-59497-3. Sci Rep. 2024. PMID: 38750104 Free PMC article.
References
- Wöhler, F. (1828) Ann. Phys. Chem. 88, 253–256.
- Sekiya, T., Takeya, T., Brown, E. L., Belagaje, R., Contreras, R., Fritz, H. J., Gait, M. J., Lees, R. G., Ryan, M. J., Khorana, H. G., et al. (1979) J. Biol. Chem. 254, 5787–5801. - PubMed
- Letsinger, R. L. & Mahadevan, V. (1965) J. Am. Chem. Soc. 87, 3526–3527. - PubMed
- Letsinger, R. L., Ogillvie, K. K. & Miller, P. S. (1969) J. Am. Chem. Soc. 91, 3360–3365.
- Matteucci, M. D. & Caruthers, M. H. (1981) J. Am. Chem. Soc. 103, 3185–3191.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials