Characterization of N protein self-association in coronavirus ribonucleoprotein complexes - PubMed (original) (raw)
Characterization of N protein self-association in coronavirus ribonucleoprotein complexes
Krishna Narayanan et al. Virus Res. 2003 Dec.
Abstract
Mouse hepatitis virus (MHV) nucleocapsid (N) protein binds to the large, single-stranded, positive-sense viral genomic RNA to form a helical nucleocapsid structure in mature virions. In addition N protein binds the intracellular form of the genomic RNA, all of the MHV subgenomic mRNAs, and expressed non-MHV RNA transcripts to form ribonucleoprotein (RNP) complexes in infected cells. Among the intracellular viral RNP complexes, only the genomic RNP complex is packaged into virus particles. The present study demonstrated that N protein in the MHV virion nucleocapsid and in the intracellular genome-length RNP complex that bound to viral envelope M protein was tightly self-associated such that its association was retained even after extensive RNase A-treatment of the RNP complexes. The RNase A-resistant tight N protein association in the virion nucleocapsid was not mediated by an intermolecular disulfide bridge between N proteins. In contrast, N protein association in the majority of the intracellular RNP complexes was susceptible to RNase A-treatment. Because the RNP complexes that specifically interact with the M protein are selectively packaged into MHV particles, the present data suggested that there was a distinct difference between N protein association in viral genomic RNP complexes that undergo packaging into virus particles and the subgenomic RNP complexes that are not packaged into MHV particles.
Figures
Fig. 1
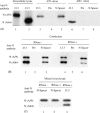
Characterization of a homotypic N protein interaction in virion nucleocapsid. (A) Purified MHV-A59 or Alb4 were disrupted by high-salt buffer, treated with RNase A and then immunoprecipitated with anti-N protein monoclonal antibody J.3.3 (lanes 3, 6), preimmune serum (lanes 4, 7), or anti-N-spacer antibody (lanes 5, 8). Radiolabeled intracellular extracts from MHV-A59-infected cells (lane 1) and Alb4-infected cells (lane 2) were also immunoprecipitated with anti-N protein monoclonal antibody J3.3. The immunoprecipitated samples were separated by SDS–PAGE. (B) Purified MHV from the cells coinfected with MHV-A59 and Alb4 were disrupted by high-salt buffer, and then treated with RNase A (lanes 4–6) or mock-treated (lanes 1–3). The samples were separated by SDS–PAGE after immunoprecipitation with anti-N protein monoclonal antibody J3.3. (lanes 1, 4), preimmune serum (lanes 2, 5), or anti-N-spacer antibody (lanes 3, 6). (C) Purified MHV-A59 and Alb4 were independently lysed with high-salt buffer, and then each virion lysate containing a similar level of N protein was mixed and treated with RNase A (lane 2, 3) or mock-treated (lane 1). The samples were then immunoprecipitated with anti-N-spacer antibody (lanes 1, 2) or J3.3 (lane 3).
Fig. 2
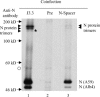
Characterization of a homotypic N protein interaction in virion under non-reducing conditions. Purified MHV from the cells coinfected with MHV-A59 and Alb4 was disrupted by high-salt buffer and then treated with RNase A. The samples were separated by non-reducing SDS–PAGE after immunoprecipitation with anti-N protein monoclonal antibody J3.3. (lane 1), preimmune serum (lane 2), or anti-N-spacer antibody (lane 3). The arrowheads indicate the N protein trimers. The asterisk represents the putative Alb4 N protein trimer. The arrows represent the positions of the 14C-labeled marker protein bands. The origin of the band, marked by open circle, is unknown.
Fig. 3
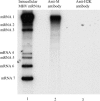
Characterization of a homotypic N protein interaction in intracellular RNP complexes. (A) Cell extracts were prepared using high-salt buffer from MHV-A59-infected cells (lanes 1, 4, 5), Alb4-infected cells (lanes 2, 6, 7) and coinfected cells (lanes 3, 8, 9). The samples were separated by SDS–PAGE after immunoprecipitation with anti-N protein monoclonal antibody J3.3. (lanes 1–3), preimmune serum (lanes 4, 6, 8), or anti-N-spacer antibody (lanes 5, 7, 9). (B) Essentially the same experimental methods were used as in (A), except that cell extracts were treated with RNase A prior to immunoprecipitation.
Fig. 4
Separation of the intracellular genomic RNP complex from other MHV intracellular subgenomic RNP complexes. 32P-radiolabeled cell extracts from MHV-A59-infected cells were prepared using the lysis buffer. The samples were incubated with either anti-M protein monoclonal antibody, J1.3 (lane 2) or anti-H2K antibody (lane 3). After immunoprecipitation, the sample was suspended in the high-salt buffer to release the coimmunoprecipitated intracellular genomic RNP complex from the antigen-antibody complex. The antigen-antibody complex was removed by centrifugation. RNA was extracted from the supernatant and examined by agarose-formaldehyde gel electrophoresis. Lane 1 represents 32P-labeled MHV-A59 mRNAs.
Fig. 5
Characterization of a homotypic N protein interaction in an intracellular genomic RNP complex. Coinfected cells were radiolabeled and cell extracts were prepared using lysis buffer. Intracellular genomic RNP complex was immunoprecipitated with anti-M monoclonal antibody, and then released from the antigen-antibody complexes by high-salt buffer treatment of the immunoprecipitates. One-half of the intracellular genomic RNP complex was incubated with RNase A (lanes 4–6), while the other half was mock-treated (lanes 1–3). After RNase treatment, the samples were immunoprecipitated with anti-N protein monoclonal antibody J3.3. (lanes 1, 4), preimmune serum (lanes 2, 5) or anti-N-spacer antibody (lanes 3, 6), and analyzed by SDS–PAGE.
Similar articles
- Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging.
Narayanan K, Makino S. Narayanan K, et al. J Virol. 2001 Oct;75(19):9059-67. doi: 10.1128/JVI.75.19.9059-9067.2001. J Virol. 2001. PMID: 11533169 Free PMC article. - Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells.
Narayanan K, Maeda A, Maeda J, Makino S. Narayanan K, et al. J Virol. 2000 Sep;74(17):8127-34. doi: 10.1128/jvi.74.17.8127-8134.2000. J Virol. 2000. PMID: 10933723 Free PMC article. - Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein.
Kuo L, Koetzner CA, Hurst KR, Masters PS. Kuo L, et al. J Virol. 2014 Apr;88(8):4451-65. doi: 10.1128/JVI.03866-13. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501403 Free PMC article. - Coronavirus genomic RNA packaging.
Masters PS. Masters PS. Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review. - Structure and assembly of the influenza A virus ribonucleoprotein complex.
Zheng W, Tao YJ. Zheng W, et al. FEBS Lett. 2013 Apr 17;587(8):1206-14. doi: 10.1016/j.febslet.2013.02.048. Epub 2013 Mar 13. FEBS Lett. 2013. PMID: 23499938 Review.
Cited by
- The nucleocapsid protein of human coronavirus NL63.
Zuwała K, Golda A, Kabala W, Burmistrz M, Zdzalik M, Nowak P, Kedracka-Krok S, Zarebski M, Dobrucki J, Florek D, Zeglen S, Wojarski J, Potempa J, Dubin G, Pyrc K. Zuwała K, et al. PLoS One. 2015 Feb 20;10(2):e0117833. doi: 10.1371/journal.pone.0117833. eCollection 2015. PLoS One. 2015. PMID: 25700263 Free PMC article. - Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers.
Cong Y, Kriegenburg F, de Haan CAM, Reggiori F. Cong Y, et al. Sci Rep. 2017 Jul 18;7(1):5740. doi: 10.1038/s41598-017-06062-w. Sci Rep. 2017. PMID: 28720894 Free PMC article. - Molecular interactions in the assembly of coronaviruses.
de Haan CA, Rottier PJ. de Haan CA, et al. Adv Virus Res. 2005;64:165-230. doi: 10.1016/S0065-3527(05)64006-7. Adv Virus Res. 2005. PMID: 16139595 Free PMC article. - Indian Ethnomedicinal Phytochemicals as Promising Inhibitors of RNA-Binding Domain of SARS-CoV-2 Nucleocapsid Phosphoprotein: An In Silico Study.
Muthumanickam S, Kamaladevi A, Boomi P, Gowrishankar S, Pandian SK. Muthumanickam S, et al. Front Mol Biosci. 2021 Jul 2;8:637329. doi: 10.3389/fmolb.2021.637329. eCollection 2021. Front Mol Biosci. 2021. PMID: 34277698 Free PMC article. - A reverse vaccinology and immunoinformatics approach for designing a multiepitope vaccine against SARS-CoV-2.
Jahangirian E, Jamal GA, Nouroozi M, Mohammadpour A. Jahangirian E, et al. Immunogenetics. 2021 Dec;73(6):459-477. doi: 10.1007/s00251-021-01228-3. Epub 2021 Sep 20. Immunogenetics. 2021. PMID: 34542663 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources