Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon - PubMed (original) (raw)
Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon
D S Nielsen et al. Appl Environ Microbiol. 2003 Dec.
Abstract
The distribution of mucosa-associated bacteria, bifidobacteria and lactobacilli and closely related lactic acid bacteria, in biopsy samples from the ascending, transverse, and descending parts of the colon from four individuals was investigated by denaturing gradient gel electrophoresis (DGGE). Bifidobacterial genus-specific, Lactobacillus group-specific, and universal bacterial primers were used in a nested PCR approach to amplify a fragment of the 16S rRNA gene. DGGE profiles of the bifidobacterial community were relatively simple, with one or two amplicons detected at most sampling sites in the colon. DGGE profiles obtained with Lactobacillus group-specific primers were complex and varied with host and sampling site in the colon. The overall bacterial community varied with host but not sampling site.
Figures
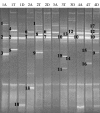
FIG. 1.
DGGE profiles representing 16S rRNA gene fragments of Bifidobacterium spp. from the ascending (A), transcending (T), and descending (D) parts of the colon wall from individuals 1 to 4. The closest relatives of the fragments sequenced (percentages of identical nucleotides compared to the sequences retrieved from the GenBank database) are as follows: B. adolescentis and B. ruminantium (1; 96.5%), B. longum and B. infantis (2; 99.4%), B. adolescentis and B. ruminantium (3; 99.4%), and B. bifidum (4; 100%).
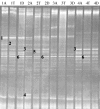
FIG. 2.
DGGE profiles representing 16S rRNA gene fragments of Lactobacillus spp. and related LAB from the ascending (A), transcending (T), and descending (D) parts of the colon wall from individuals 1 to 4. The closest relatives of fragments sequenced (percentages of identical nucleotides compared to sequences retrieved from the GenBank database) are as follows: Lactobacillus fermentum (1; 94.5%), E. biforme (2; 99.5%); Leuconostoc carnosum (3; 97.8%); Aerococcus viridans (4; 97.2%); Aerococcus sanguinicola (5; 97.3%); Weissella kimchii, Weissella cibaria, and Weissella confusa (6 and 7; 99.5 and 98.0%, respectively); L. fermentum (8; 96.8%); Leuconostoc citreum (9; 98.9%); L. citreum (10; 98.9%); L. citreum (11; 94.6%); Leuconostoc kimchii (12; 98.8%); Leuconostoc fallax (13; 99.5%); Lactobacillus iners (14; 99.4%); Weissella soli (15; 95.1%); L. ruminis (16; 96.2%); W. soli (17; 95.1%). 18, no sequence obtained or the fragment did not migrate as an original band after reamplification.
FIG. 3.
Dendrogram derived from DGGE analysis of the _Lactobacillus_-like population from the ascending (A), transcending (T), and descending (D) parts of the colon wall from individuals 1 to 4 based on Dice's coefficient of similarity with the unweighted pair group method with arithmetic averages clustering algorithm.
FIG. 4.
DGGE profiles representing 16S rRNA gene fragments of the predominating bacteria from the ascending (A), transcending (T), and descending (D) parts of the colon wall from individuals 1 to 4. Closest relatives of fragments sequenced (percentages of identical nucleotides compared to sequence retrieved from the GenBank database) are as follows: Bacteroides vulgatus (1; 95.0%), Bacteroides vulgatus (2; 95.6%); Prevotella enoeca (3; 91.7%); B. bifidum (4; 100%); Bacteroides distasonis (5; 96.1%). 6, no sequence obtained.
References
- Boon, N., W. de Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. - PubMed
- Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. - PMC - PubMed
- Hentschel, U., U. Dobrindt, and M. Steinert. 2003. Commensal bacteria make a difference. Trends Microbiol. 11:148-150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources