Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management - PubMed (original) (raw)
Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management
Dag Harmsen et al. J Clin Microbiol. 2003 Dec.
Abstract
The spa gene of Staphylococcus aureus encodes protein A and is used for typing of methicillin-resistant Staphylococcus aureus (MRSA). We used sequence typing of the spa gene repeat region to study the epidemiology of MRSA at a German university hospital. One hundred seven and 84 strains were studied during two periods of 10 and 4 months, respectively. Repeats and spa types were determined by Ridom StaphType, a novel software tool allowing rapid repeat determination, data management and retrieval, and Internet-based assignment of new spa types following automatic quality control of DNA sequence chromatograms. Isolates representative of the most abundant spa types were subjected to multilocus sequence typing and pulsed-field gel electrophoresis. One of two predominant spa types was replaced by a clonally related variant in the second study period. Ten unique spa types, which were equally distributed in both study periods, were recovered. The data show a rapid dynamics of clone circulation in a university hospital setting. spa typing was valuable for tracking of epidemic isolates. The data show that disproval of epidemiologically suggested transmissions of MRSA is one of the main objectives of spa typing in departments with a high incidence of MRSA.
Figures
FIG. 1.
Screen shot of the novel Ridom StaphType software featuring a base quality-based sequence editor, a database, and a report generator (not shown) module. This client software synchronizes with an accompanying website to ensure uniform spa code terminology usage.
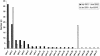
FIG. 2.
Rank abundance graph demonstrating the frequencies of spa types collected during two study periods.
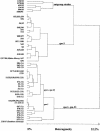
FIG. 3.
Dendrogram deduced from the cluster analysis of PFGE macrorestriction patterns of spa-1, -3, and -23 strains as determined by the algorithm described by Claus et al. (1). Strains from this study are designated by the letter S followed by a strain number. Additional information was added to spa-1 and -23 strains as follows: 1s, spa-1 and low mupirocin MIC; 1i, spa-1 and high mupirocin MIC; 23i, spa-23 and high mupirocin MIC. As a control, patterns determined previously from diverse other clones at the German reference laboratory for staphylococci were included for cluster analysis (outgroup strains, i.e., 1150/93, Berlin epidemic MRSA; 2378/00, Barnim epidemic MRSA; 635/93, Vienna epidemic MRSA; 134/93, northern German epidemic MRSA; 1000/93, Hannover epidemic MRSA).
Similar articles
- [Spa types and antibiotic resistance of Staphylococcus aureus bloodstream isolates obtained form patients of the University Clinical Center in Gdańsk].
Wiśniewska K, Kasprzyk J, Piechowicz L, Bronk M, Swieć K. Wiśniewska K, et al. Med Dosw Mikrobiol. 2013;65(3):139-47. Med Dosw Mikrobiol. 2013. PMID: 24432553 Polish. - [Evidence-based infection control methods using spa genotyping for MRSA spread in hospitals].
Mellmann A, Friedrich AW, Kipp F, Hinder F, Keckevoet U, Harmsen D. Mellmann A, et al. Dtsch Med Wochenschr. 2005 Jun 3;130(22):1364-8. doi: 10.1055/s-2005-868735. Dtsch Med Wochenschr. 2005. PMID: 15915378 German. - DNA sequence-based tandem repeat analysis of the clfB gene is less discriminatory than spa typing for methicillin-resistant Staphylococcus aureus.
Harmsen D, Claus H, Vogel U. Harmsen D, et al. Int J Med Microbiol. 2005 Mar;294(8):525-8. doi: 10.1016/j.ijmm.2004.11.002. Int J Med Microbiol. 2005. PMID: 15790297 - The evolution of Staphylococcus aureus.
Deurenberg RH, Stobberingh EE. Deurenberg RH, et al. Infect Genet Evol. 2008 Dec;8(6):747-63. doi: 10.1016/j.meegid.2008.07.007. Epub 2008 Jul 29. Infect Genet Evol. 2008. PMID: 18718557 Review. - Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus.
Robinson DA, Enright MC. Robinson DA, et al. Clin Microbiol Infect. 2004 Feb;10(2):92-7. doi: 10.1111/j.1469-0691.2004.00768.x. Clin Microbiol Infect. 2004. PMID: 14759234 Review.
Cited by
- Transcriptional Profiling of Staphylococcus aureus during the Transition from Asymptomatic Nasal Colonization to Skin Colonization/Infection in Patients with Atopic Dermatitis.
Li P, Schulte J, Wurpts G, Hornef MW, Wolz C, Yazdi AS, Burian M. Li P, et al. Int J Mol Sci. 2024 Aug 23;25(17):9165. doi: 10.3390/ijms25179165. Int J Mol Sci. 2024. PMID: 39273114 Free PMC article. - The circulation of methicillin-resistant Staphylococcus aureus between humans, horses and the environment at the equine clinic.
Papouskova A, Drabkova Z, Brajerova M, Krutova M, Cizek A, Tkadlec J. Papouskova A, et al. J Antimicrob Chemother. 2024 Nov 4;79(11):2901-2905. doi: 10.1093/jac/dkae303. J Antimicrob Chemother. 2024. PMID: 39212167 Free PMC article. - Genetic Characterization of Antibiotic-Resistant Staphylococcus spp. and Mammaliicoccus sciuri from Healthy Humans and Poultry in Nigeria.
Jesumirhewe C, Odufuye TO, Ariri JU, Adebiyi AA, Sanusi AT, Stöger A, Daza-Prieto B, Allerberger F, Cabal-Rosel A, Ruppitsch W. Jesumirhewe C, et al. Antibiotics (Basel). 2024 Aug 5;13(8):733. doi: 10.3390/antibiotics13080733. Antibiotics (Basel). 2024. PMID: 39200033 Free PMC article. - Molecular characteristics and antimicrobial resistance profiles of Staphylococcus aureus isolates from burns.
Kadhum DA, Hamad EM, Fahad MA. Kadhum DA, et al. Mol Biol Rep. 2024 Aug 12;51(1):903. doi: 10.1007/s11033-024-09837-3. Mol Biol Rep. 2024. PMID: 39133365 - Anti-virulence potential of iclaprim, a novel folic acid synthesis inhibitor, against Staphylococcus aureus.
Hao L, Zhou J, Yang H, He C, Shu W, Song H, Liu Q. Hao L, et al. Appl Microbiol Biotechnol. 2024 Aug 5;108(1):432. doi: 10.1007/s00253-024-13268-2. Appl Microbiol Biotechnol. 2024. PMID: 39102054 Free PMC article.
References
- Claus, H., C. Cuny, B. Pasemann, and W. Witte. 1998. A database system for fragment patterns of genomic DNA of Staphylococcus aureus. Zentbl. Bakteriol. 287:105-116. - PubMed
- Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. - PubMed
- Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources