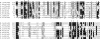Trans-editing of mischarged tRNAs - PubMed (original) (raw)
Trans-editing of mischarged tRNAs
Ivan Ahel et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Aminoacyl-tRNA synthetases (aaRSs) are multidomain proteins that specifically attach amino acids to their cognate tRNAs. Their most conserved, and presumably evolutionarily oldest, domains are the catalytic cores, which activate amino acids and transfer them to the 3' ends of tRNAs. Additional domains appended to or inserted in the body of aaRSs increase efficiency and specificity of the aminoacylation process, either by providing additional tRNA contacts, or by hydrolyzing noncognate amino acid products (cis-editing). Here, we report specific tRNA-dependent trans-editing by aaRS-like proteins that reciprocate the editing domains of aaRSs, but not the remainder of the corresponding enzyme. A freestanding homologue of the prolyl-tRNA synthetase-editing domain, the PrdX protein from Clostridium sticklandii, efficiently and specifically hydrolyzes Ala-tRNAPro. Similarly, autonomous alanyl-tRNA synthetase-editing domain homologues (AlaX proteins) from Methanosarcina barkeri and Sulfolobus solfataricus hydrolyze Ser-tRNAAla and Gly-tRNAAla substrates. The discovery of autonomous editing proteins efficient in hydrolyzing misacylated products provides a direct link between ancestral aaRSs consisting solely of the catalytic core and extant enzymes to which functionally independent modules are appended.
Figures
Fig. 1.
Multiple alignment of the ProRS editing domain homologues. Asterisks represent residues that were shown important for editing function of the E. coli ProRS enzyme. “>OXH<” defines the position of the oxyanion hole. The last two sequences represent Ybak proteins [the crystal structure has been solved for the H. influenzae protein (17)]. M.mus, Mus musculus (NM_026465); M.mag, Magnetospirillum magnetotacticum (ZP_00053959); R.rub, Rhodospirillum rubrum (ZP_00016210); C.sti, C. sticklandii (CAB71308); P.fal, P. falciparum (AE014846); T.bru, Trypanosoma brucei (
); E.col, E. coli (ProRS AAB08622, Ybak AAC73583); B.sub, Bacillus subtilis (NP_389539); C.ace, Clostridium acetobutylicum (NP_349775); A.aeo, Aquifex aeolicus (NP_213250); H.inf, H. influenzae (P45202); Ins, insertion domain of bacterial-type ProRS; Nterm, N-terminal extension in ProRSs from lower eukaryotes. GenBank accession numbers are in parentheses.
Fig. 2.
Different architectures of the Prolyl-tRNA synthetases and their editing domains. In blue are shown editing domains (note that they can be present in cis or trans; as an N-terminal domain or an insertion). In red are shown catalytic domains (orange is used for bacterial-type ProRS). Anticodon-binding domains are yellow, and archaeal-type ProRS-specific C termini are green. Representatives of the analyzed editing domain homologues are shown in parentheses.
Fig. 3.
Unrooted phylogenetic tree implied by the neighbor-joining method showing possible relationships between ProRS insertion domain homologues. If not specifically labeled in genomic databases, the free-standing ProRS editing domain homologues are denoted ProX. GenBank accession numbers are in parentheses: C. sticklandii PrdX (CAB71308), Pseudomonas aeruginosa ProX (AAG05230), M. magnetotacticum ProX (ZP_00053959), Agrobacterium tumefaciens ProX1 (NP_356919), M. musculus ProX (NM_026465), Arabidopsis thaliana ProX (AAO63892), P. falciparum ProRS (AE014846), Chlamydia trachomatis ProRS (NP_219903), E. coli ProRS (AAB08622), Thermotoga maritima ProRS (NP_228324), C. acetobutylicum ProRS (NP_349775), E. coli YeaK (AAC74857), Chlorobium tepidum ProX (NP_662760), Aeropyrum pernix ProX (NP_148681), A. tumefaciens ProX2 (AAK85924), Enterococcus faecalis EbsC (AAC36853), Streptomyces coelicolor ProX (NP_626318), E. coli YbaK (AAC73583), and H. influenzae YbaK (P45202).
Fig. 4.
Multiple alignments of the editing modules of AlaRS and ThrRS. Asterisks represent residues that were shown to be important for the editing function of the E. coli ThrRS or the E. coli AlaRS enzymes. M.jan, M. jannaschii (Q57984); P.aby, Pyrococcus abyssi (Q9UY36); E.col, E. coli (Q8XE27); B.sub, B. subtilis (P18256); M.bar, M. barkeri (ZP_00076591); A.per, A. pernix (NP_147589); S.sol, S. solfataricus (AlaX1, NP_341986; AlaX2, NP_342718); D.rad, Deinococcus radiodurans (NP_294225). GenBank accession numbers are in parentheses. S. solfataricus possesses two different AlaX.
Fig. 5.
Unrooted phylogenetic tree implied by the neighbor-joining method, showing relationships between the editing domains of AlaRS and ThrRS and their free-standing homologues found elsewhere. Note that the homologous module is found as an insert (AlaRS), an N-terminal extension (ThrRS), and a freestanding protein (AlaX). A homologous domain was also found at the N terminus of Plasmodium TrpRS. GenBank accession numbers are in parentheses: D. radiodurans AlaX (NP_294225), M. barkeri AlaX (ZP_00076591), S. solfataricus AlaX1 (NP_341986), P. aeruginosa AlaX (NP_250796), Mesorhizobium loti AlaX (NP_102167), D. radiodurans AlaRS (Q9RS27), C. acetobutylicum AlaRS (Q97IG3), Giardia lamblia AlaRS (AAG23137), M. barkeri AlaRS (ZP_00077929), M. jannaschii AlaRS (Q57984), Thermoplasma volcanium ThrRS (Q978W0), Bacillus halodurans ThrRS (Q9K866), T. maritima ThrRS (Q9WZJ9), Rickettsia prowazekii ThrRS (O05947), E. coli ThrRS (Q8XE27), P. falciparum TrpRS (NP_705269), Dictyostelium discoideum AlaX (AAO52108), Pyrococcus horikoshii AlaX2 (NP_142539), S. solfataricus AlaX2 (NP_342718), Homo sapiens AlaX (NP_079543), S. cerevisiae AlaX (NP_014358).
Fig. 6.
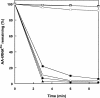
(A) Specific deacylation of E. coli Ala-tRNAPro. Shown are 0.3 μM CSPrdX (•), 0.3 μM CSPrdX plus 1 μM CSProRS (▴), 0.3 μM ECYbaK (▪), 0.5 μM PFProRS (○), 0.5 μM ΔPFProRS (□), 0.5 μM S. cerevisiae ProRS (▵), 0.1 μM E. coli ProRS (▾), and control without enzyme (▿). (Inset) Pro-tRNAPro and Ala-tRNAAla deacylation [0.3 μM CSPrdX (• and ○, respectively), 0.5 μM PFProRS (▪ and □, respectively)]. (B) Mischarging of alanine onto S. cerevisiae tRNAPro. Shown are 6 μM PFProRS (•), 6 μM ΔPFProRS (▪), and 6 μM S. cerevisiae ProRS (▵).
Fig. 7.
Specific deacylations by AlaXs. Shown are 100 nM M. barkeri AlaX and Ser-tRNAAla (•), 100 nM S. solfataricus AlaX and Ser-tRNAAla (▪), 100 nM M. jannaschii AlaRS and Ser-tRNAAla (▴), 100 nM S. cerevisiae AlaX and Ser-tRNAAla (□), 100 nM M. barkeri AlaX and Gly-tRNAAla (○), 100 nM M. barkeri AlaX and Ser-tRNAThr (▾), and control without enzyme (▵).
Similar articles
- Human cytoplasmic ProX edits mischarged tRNAPro with amino acid but not tRNA specificity.
Ruan LL, Zhou XL, Tan M, Wang ED. Ruan LL, et al. Biochem J. 2013 Feb 15;450(1):243-52. doi: 10.1042/BJ20121493. Biochem J. 2013. PMID: 23210460 - Ancestral AlaX editing enzymes for control of genetic code fidelity are not tRNA-specific.
Novoa EM, Vargas-Rodriguez O, Lange S, Goto Y, Suga H, Musier-Forsyth K, Ribas de Pouplana L. Novoa EM, et al. J Biol Chem. 2015 Apr 17;290(16):10495-503. doi: 10.1074/jbc.M115.640060. Epub 2015 Feb 27. J Biol Chem. 2015. PMID: 25724653 Free PMC article. - Trans-editing by aminoacyl-tRNA synthetase-like editing domains.
Kuzmishin Nagy AB, Bakhtina M, Musier-Forsyth K. Kuzmishin Nagy AB, et al. Enzymes. 2020;48:69-115. doi: 10.1016/bs.enz.2020.07.002. Epub 2020 Sep 8. Enzymes. 2020. PMID: 33837712 Review. - A new mechanism of post-transfer editing by aminoacyl-tRNA synthetases: catalysis of hydrolytic reaction by bacterial-type prolyl-tRNA synthetase.
Boyarshin KS, Priss AE, Rayevskiy AV, Ilchenko MM, Dubey IY, Kriklivyi IA, Yaremchuk AD, Tukalo MA. Boyarshin KS, et al. J Biomol Struct Dyn. 2017 Feb;35(3):669-682. doi: 10.1080/07391102.2016.1155171. Epub 2016 Mar 8. J Biomol Struct Dyn. 2017. PMID: 26886480 - Quality control in tRNA charging.
Jakubowski H. Jakubowski H. Wiley Interdiscip Rev RNA. 2012 May-Jun;3(3):295-310. doi: 10.1002/wrna.122. Epub 2011 Nov 17. Wiley Interdiscip Rev RNA. 2012. PMID: 22095844 Review.
Cited by
- Substrate specificity of bacterial prolyl-tRNA synthetase editing domain is controlled by a tunable hydrophobic pocket.
Kumar S, Das M, Hadad CM, Musier-Forsyth K. Kumar S, et al. J Biol Chem. 2012 Jan 27;287(5):3175-84. doi: 10.1074/jbc.M111.313619. Epub 2011 Nov 29. J Biol Chem. 2012. PMID: 22128149 Free PMC article. - An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase.
Khan S, Garg A, Sharma A, Camacho N, Picchioni D, Saint-Léger A, Ribas de Pouplana L, Yogavel M, Sharma A. Khan S, et al. PLoS One. 2013 Jun 12;8(6):e66224. doi: 10.1371/journal.pone.0066224. Print 2013. PLoS One. 2013. PMID: 23776638 Free PMC article. - Cellular mechanisms that control mistranslation.
Reynolds NM, Lazazzera BA, Ibba M. Reynolds NM, et al. Nat Rev Microbiol. 2010 Dec;8(12):849-56. doi: 10.1038/nrmicro2472. Nat Rev Microbiol. 2010. PMID: 21079633 Review. - Genomic innovation of ATD alleviates mistranslation associated with multicellularity in Animalia.
Kuncha SK, Venkadasamy VL, Amudhan G, Dahate P, Kola SR, Pottabathini S, Kruparani SP, Shekar PC, Sankaranarayanan R. Kuncha SK, et al. Elife. 2020 May 28;9:e58118. doi: 10.7554/eLife.58118. Elife. 2020. PMID: 32463355 Free PMC article. - Aminoacyl-tRNA synthetases as drug targets in eukaryotic parasites.
Pham JS, Dawson KL, Jackson KE, Lim EE, Pasaje CF, Turner KE, Ralph SA. Pham JS, et al. Int J Parasitol Drugs Drug Resist. 2013 Nov 11;4(1):1-13. doi: 10.1016/j.ijpddr.2013.10.001. eCollection 2014 Apr. Int J Parasitol Drugs Drug Resist. 2013. PMID: 24596663 Free PMC article. Review.
References
- Fersht, A. R. (1981) Proc. R. Soc. London Ser. B. 212, 351–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous