Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis - PubMed (original) (raw)
Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis
Eric Jan et al. Proc Natl Acad Sci U S A. 2003.
Abstract
The cricket paralysis virus internal ribosome entry site (IRES) can, in the absence of canonical initiation factors and initiator tRNA (Met-tRNAi), occupy the ribosomal P-site and assemble 80S ribosomes. Here we show that the IRES assembles mRNA-80S ribosome complexes by recruitment of 60S subunits to preformed IRES-40S complexes. Addition of eukaryotic elongation factors eEF1A and eEF2 and aminoacylated elongator tRNAs resulted in the synthesis of peptides, implying that the IRES RNA itself mimics the function of Met-tRNAi in the P-site to trigger the first translocation step without peptide bond formation. IRES-80S complexes that contained a stop codon in the A-site recruited eukaryotic release factor eRF1, resulting in ribosome rearrangements in a surprisingly eEF2-dependent manner. Thus, this P-site-occupying IRES directs the assembly of 80S ribosomes, sets the translational reading frame, and mimics the functions of both Met-tRNAi and peptidyl tRNA to support elongation and termination.
Figures
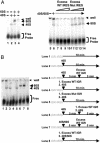
Fig. 1.
80S ribosome assembly on the IGR IRES using purified 40S and 60S subunits. (A)5′ end-labeled IGR IRES RNAs (0.5 nM) were incubated alone (lane 1), with 40S (14 nM, lane 2), with 60S (16 nM, lane 4), or with 40S and 60S (lane 3) ribosomal subunits in 5-μl reactions. Excess (150 nM, 300 nM, 600 nM, and 800 nM) unlabeled WT IGR IRES RNAs (lanes 7–10) and mutated (Mut) IRES RNAs (lanes 11–14) were added as indicated. Excess RNAs were added to reactions before addition of 40S and 60S subunits. Reactions were separated on composite agarose polyacrylamide gels, dried, and exposed to x-ray film. (B) Formation of IRES-80S complexes by joining 60S to preformed binary IRES-40S complexes. (Right) The sequential order of reagents is shown. (Left) Radiolabeled WT IGR IRES RNAs were incubated alone (lane 1) or with 40S subunits (lane 2). Excess unlabeled WT IGR IRES RNAs or mutated (Mut) IGR IRES RNAs were added after (lane 3) or before (lanes 4 and 5) 40S subunit incubation. Addition of 60S subunits to prebound WT IGR IRES-40S complexes assembled 80S complexes in the presence of excess unlabeled WT IGR IRES RNAs (lane 6). Excess unlabeled WT IGR IRES RNAs were added after (lane 7) or before (lane 8) addition of 40S and 60S subunits. Reactions were separated on composite agarose polyacrylamide gels, dried, and exposed to x-ray film.
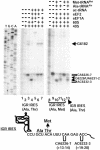
Fig. 2.
Peptide synthesis directed by the IGR IRES in a reconstituted system. Dicistronic constructs containing the IGR IRES were engineered to synthesize ≈3-kDa (IGR-ORFS) or 6.5-kDa (IGR-ORFL) peptides. RNAs were incubated with 40S and 60S subunits for 5 min at 37°C, followed by addition of elongation factors and aminoacylated tRNAs and further incubation for 25 min at 37°C. Translation products from WT or mutated (Mut) IRES-containing RNAs are shown as indicated. Reactions were treated with 0.7 mg/ml RNase A (lane 2) or 2 mg/ml proteinase (lane 3) (Pronase, Boehringer Mannheim). Cycloheximide (Cyclohex., 500 μg/ml, lane 4) was added to the reactions before the addition of elongation factors. Mutated IGR IRES (Mut-ORFS) contains the mutation CC6214-5GG, which disrupts pseudoknot I (6). Lane 9, reaction containing the EMCV IRES (EMCV-ORFS) is shown. All reactions were loaded on 16.5% Tris·Tricine gels, which were dried and exposed to x-ray films. Autoradiographs are shown.
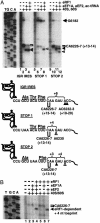
Fig. 3.
Ribosome rearrangement in IGR IRES–ribosome complexes in a reconstituted system. Movement of the ribosome on the IGR IRES was monitored by toeprinting analysis. Ribosomes assembled on dicistronic RNAs containing the WT IGR IRES (Ala, Thr) (lanes 2–7 and 13) or a WT IGR IRES (Ala, Met) with a Ala GCU codon followed by a Met AUG codon (lanes 8–12). Purified 40S, 60S, eEF2, eEF1A, and bulk aminoacylated tRNAs (ac-tRNA) were added in combinations to reactions with the dicistronic RNAs and analyzed by primer extension analysis (lanes 1–7). All reactions shown were in the presence of cycloheximide. Ala-tRNAs synthesized in vitro and elongator Met-tRNAs were used in lanes 8–12 instead of bulk aminoacylated tRNAs. Reactions were displayed on denaturing polyacrylamide gels and quantitated by PhosphorImager analysis. Toeprints of ribosome translocation on the IGR IRES are marked on the right. Numbering of the nucleotides refers to the nucleotide's position in the CrPV genome. A summary of the toeprints on the dicistronic RNAs is shown below the gel.
Fig. 4.
P-site-located IGR IRES modulates the movement of A-site-located stop codon–eRF1 complexes. (A) Ribosome translocation on a dicistronic RNA containing the WT IGR IRES or the WT IGR IRES with a UAA stop codon in the first (STOP 1) or second (STOP 2) triplet of the downstream ORF. IGR IRES, STOP 1, or STOP 2 dicistronic RNAs were incubated alone (lanes 1, 5, and 9); with 40S and 60S subunits (lanes 2, 6, and 10); with 40S, 60S, elongation factors, and aminoacylated tRNAs (lanes 3, 7, and 11); or with 40S, 60S, elongation factors, aminoacylated tRNAs, and purified eRF1 (lanes 4, 8, and 12). All reactions were incubated in the presence of cycloheximide. Reactions were analyzed by toeprinting assays, displayed on denaturing polyacrylamide gels, and quantitated by PhosphorImager analysis. A summary of the toeprinting analyses is shown below the gel. Brackets denote the location of toeprints induced by ribosome that had not translocated. Filled arrowheads point to toeprints of ribosomes that had translocated. Open arrowheads indicate the +4-nt toeprint observed in the presence of eRF1. (B) eRF1-induced toeprint depends on the presence of eEF2. STOP 1 dicistronic RNAs were incubated alone (lane 1), with 40S and 60S subunits (lane 2), with 40S, 60S, eEF1A, and eEF2 (lane 3), or with 40S, 60S, eEF1A, eEF2, and eRF1 (lane 4). eEF1A, eEF2, and eRF1 were each omitted from the reaction (lanes 5–7). Reactions were analyzed by toeprinting assay and displayed on a denaturing polyacrylamide gel as above.
Similar articles
- Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site.
Lancaster AM, Jan E, Sarnow P. Lancaster AM, et al. RNA. 2006 May;12(5):894-902. doi: 10.1261/rna.2342306. Epub 2006 Mar 23. RNA. 2006. PMID: 16556939 Free PMC article. - Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites.
Pisarev AV, Shirokikh NE, Hellen CU. Pisarev AV, et al. C R Biol. 2005 Jul;328(7):589-605. doi: 10.1016/j.crvi.2005.02.004. C R Biol. 2005. PMID: 15992743 Review. - Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES.
Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. Muhs M, et al. Mol Cell. 2015 Feb 5;57(3):422-32. doi: 10.1016/j.molcel.2014.12.016. Epub 2015 Jan 15. Mol Cell. 2015. PMID: 25601755 Free PMC article. - [Interaction of tRNA with ribosomes].
Kirillov SV, Semenkov IuP. Kirillov SV, et al. Mol Biol (Mosk). 1984 Sep-Oct;18(5):1249-63. Mol Biol (Mosk). 1984. PMID: 6390173 Review. Russian.
Cited by
- Modular domains of the Dicistroviridae intergenic internal ribosome entry site.
Jang CJ, Jan E. Jang CJ, et al. RNA. 2010 Jun;16(6):1182-95. doi: 10.1261/rna.2044610. Epub 2010 Apr 27. RNA. 2010. PMID: 20423979 Free PMC article. - Kinetics of initiating polypeptide elongation in an IRES-dependent system.
Zhang H, Ng MY, Chen Y, Cooperman BS. Zhang H, et al. Elife. 2016 Jun 2;5:e13429. doi: 10.7554/eLife.13429. Elife. 2016. PMID: 27253065 Free PMC article. - Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts.
Hatakeyama Y, Shibuya N, Nishiyama T, Nakashima N. Hatakeyama Y, et al. RNA. 2004 May;10(5):779-86. doi: 10.1261/rna.5208104. RNA. 2004. PMID: 15100433 Free PMC article. - tRNA-mRNA mimicry drives translation initiation from a viral IRES.
Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. Costantino DA, et al. Nat Struct Mol Biol. 2008 Jan;15(1):57-64. doi: 10.1038/nsmb1351. Epub 2007 Dec 23. Nat Struct Mol Biol. 2008. PMID: 18157151 Free PMC article. - Poliovirus switches to an eIF2-independent mode of translation during infection.
White JP, Reineke LC, Lloyd RE. White JP, et al. J Virol. 2011 Sep;85(17):8884-93. doi: 10.1128/JVI.00792-11. Epub 2011 Jun 22. J Virol. 2011. PMID: 21697471 Free PMC article.
References
- Dever, T. E. (2002) Cell 108, 545–556. - PubMed
- Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593–1612. - PubMed
- Wilson, J. E., Pestova, T. V., Hellen, C. U. & Sarnow, P. (2000) Cell 102, 511–520. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous