Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide - PubMed (original) (raw)
Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide
Stephen Tottey et al. Proc Natl Acad Sci U S A. 2003.
Abstract
CHL27, the Arabidopsis homologue to Chlamydomonas Crd1, a plastid-localized putative diiron protein, is required for the synthesis of protochlorophyllide and therefore is a candidate subunit of the aerobic cyclase in chlorophyll biosynthesis. delta-Aminolevulinic acid-fed antisense Arabidopsis plants with reduced amounts of Crd1/CHL27 accumulate Mg-protoporphyrin IX monomethyl ester, the substrate of the cyclase reaction. Mutant plants have chlorotic leaves with reduced abundance of all chlorophyll proteins. Fractionation of Arabidopsis chloroplast membranes shows that Crd1/CHL27 is equally distributed on a membrane-weight basis in the thylakoid and inner-envelope membranes.
Figures
Fig. 1.
The oxidative cyclase reaction for the formation of the fifth ring of the Chl molecule. The conversion of MgPMME to divinyl Pchlide proceeds through three sequential two-electron oxidations. The first step requires molecular oxygen as a substrate for hydroxylation, analogous to the methane monooxygenase reaction. There is also an oxygen requirement for the third reaction.
Fig. 2.
Phenotypes of _chl27_-as Arabidopsis plants. (Ai) A plant line (_chl27_-as1.7.5) that shows a severe phenotype early after transfer from sucrose-containing plates to soil. (Aii) A line (_chl27_-as2.38.10) that shows a severe chlorotic phenotype at a later stage of development. (Aiii) A plant line (_chl27_-as2.38.4) that displays a variegated phenotype. (Aiv) A WT plant transformed with pPZP111 (vector control). (B) Immunoblot detection of CHL27. Plant material was collected from chlorotic lines (1, _chl27_-as1.7.5; 2, _chl27_-as2.22.7), variegated lines (3, _chl27_-as2.22.10; 4, _chl27_-as2.22.12), green lines (5, _chl27_-as2.22.1; 6, _chl27_-as2.22.6), and WT lines (7 and 8). Proteins were resolved by SDS/PAGE, and the abundance of CHL27 was revealed by immunoblotting. Each lane contains 30 μg of total protein.
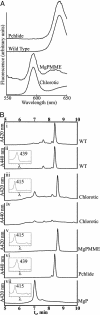
Fig. 3.
Analysis of porphyrin pigments accumulating in _chl27_-as lines after feeding with ALA in the dark. (A) Room-temperature fluorescence emission spectra of acetone extracts of chlorotic (yellow) leaves from an antisense line (_chl27_-as2.38.10) and green leaves from WT plants after overnight incubation with ALA compared to purified standards. (B) HPLC traces of the extracted pigment mixtures relative to known standards. The eluate was monitored at 420 nm for optimal detection of MgPMME and MgP (i, iii, v, and vii) or 440 nm for detection of Pchlide (ii, iv, and vi). (Inset) Traces show spectral absorbance of the major eluate peak, measured from 350 to 595 nm, with the wavelength of maximum absorbance indicated.
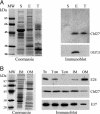
Fig. 4.
Suborganellar localization of CHL27. (A) Analysis of Arabidopsis chloroplast fractions. E, envelope; T, thylakoid; S, stroma. (Left) Fractions analyzed after SDS/PAGE by Coomassie blue staining. (Right) Immunoblot detection of CHL27 and OEP21. (B) Distribution of CHL27 between envelope fractions from spinach chloroplasts. Envelope membrane fractions were enriched in inner-envelope membrane (IM) or outer-envelope membrane (OM). T0,T100, and T600, envelope fractions were obtained from chloroplasts treated without thermolysin or with 100 or 600 μg/ml thermolysin, respectively. (Left) Coomassie blue staining. (Right) Immunodetection of CHL27, E37, an inner-envelope membrane protein, and E24, an outer-envelope membrane protein. Each lane was loaded with 20 μg of protein. The membrane was incubated with anti-CHL27 (1:1,000), anti-E37 (1:10,000), and anti-E24 (1:5,000).
Fig. 5.
Reduced Chl protein abundance in _chl27_-as plants. (A) Fluorescence emissions (77 K) measured from 650 to 800 nm from WT (black), antisense lines showing no visible phenotype [green (_chl27_-as2.22.1) and light green (_chl27_-as2.22.6)], variegated lines [blue (_chl27_-as2.22.10) and light blue (_chl27_-as2.22.12)], and chlorotic [orange (_chl27_-as1.7.5)] plants. (B) Immunoblot analysis of CHL27-deficient and WT plants. Thylakoid membranes isolated from lines 2–14 (mild variegation) and 1–7 (more severe variegation) were analyzed for the abundance of the indicated thylakoid membrane protein. Each lane was loaded with 3 μg of protein.
Similar articles
- Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling.
Bang WY, Jeong IS, Kim DW, Im CH, Ji C, Hwang SM, Kim SW, Son YS, Jeong J, Shiina T, Bahk JD. Bang WY, et al. Plant Cell Physiol. 2008 Sep;49(9):1350-63. doi: 10.1093/pcp/pcn111. Epub 2008 Aug 4. Plant Cell Physiol. 2008. PMID: 18682427 - Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves.
Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL. Wang Q, et al. Plant Physiol. 2004 Nov;136(3):3594-604. doi: 10.1104/pp.104.049841. Epub 2004 Oct 29. Plant Physiol. 2004. PMID: 15516501 Free PMC article. - The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings.
Tanz SK, Kilian J, Johnsson C, Apel K, Small I, Harter K, Wanke D, Pogson B, Albrecht V. Tanz SK, et al. Plant J. 2012 Mar;69(5):743-54. doi: 10.1111/j.1365-313X.2011.04833.x. Epub 2011 Dec 2. Plant J. 2012. PMID: 22040291 - Differential distribution of chlorophyll biosynthetic intermediates in stroma, envelope and thylakoid membranes in Beta vulgaris.
Mohapatra A, Tripathy BC. Mohapatra A, et al. Photosynth Res. 2007 Nov-Dec;94(2-3):401-10. doi: 10.1007/s11120-007-9209-6. Epub 2007 Jul 19. Photosynth Res. 2007. PMID: 17638115 - The Plastid Terminal Oxidase is a Key Factor Balancing the Redox State of Thylakoid Membrane.
Wang D, Fu A. Wang D, et al. Enzymes. 2016;40:143-171. doi: 10.1016/bs.enz.2016.09.002. Epub 2016 Oct 12. Enzymes. 2016. PMID: 27776780 Review.
Cited by
- PAPP5 is involved in the tetrapyrrole mediated plastid signalling during chloroplast development.
Barajas-López Jde D, Kremnev D, Shaikhali J, Piñas-Fernández A, Strand A. Barajas-López Jde D, et al. PLoS One. 2013;8(3):e60305. doi: 10.1371/journal.pone.0060305. Epub 2013 Mar 29. PLoS One. 2013. PMID: 23555952 Free PMC article. - Arabidopsis transcription factor TCP4 represses chlorophyll biosynthesis to prevent petal greening.
Zheng X, Lan J, Yu H, Zhang J, Zhang Y, Qin Y, Su XD, Qin G. Zheng X, et al. Plant Commun. 2022 Jul 11;3(4):100309. doi: 10.1016/j.xplc.2022.100309. Epub 2022 Mar 3. Plant Commun. 2022. PMID: 35605201 Free PMC article. - Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells.
Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ. Majeran W, et al. Mol Cell Proteomics. 2008 Sep;7(9):1609-38. doi: 10.1074/mcp.M800016-MCP200. Epub 2008 May 2. Mol Cell Proteomics. 2008. PMID: 18453340 Free PMC article. - Ferredoxin C2 is required for chlorophyll biosynthesis and accumulation of photosynthetic antennae in Arabidopsis.
Tournaire MD, Scharff LB, Kramer M, Goss T, Vuorijoki L, Rodriguez-Heredia M, Wilson S, Kruse I, Ruban A, Balk L J, Hase T, Jensen PE, Hanke GT. Tournaire MD, et al. Plant Cell Environ. 2023 Nov;46(11):3287-3304. doi: 10.1111/pce.14667. Epub 2023 Jul 10. Plant Cell Environ. 2023. PMID: 37427830 Free PMC article. - Absence of the cbb3 Terminal Oxidase Reveals an Active Oxygen-Dependent Cyclase Involved in Bacteriochlorophyll Biosynthesis in Rhodobacter sphaeroides.
Chen GE, Canniffe DP, Martin EC, Hunter CN. Chen GE, et al. J Bacteriol. 2016 Jul 13;198(15):2056-63. doi: 10.1128/JB.00121-16. Print 2016 Aug 1. J Bacteriol. 2016. PMID: 27215788 Free PMC article.
References
- Suzuki, J. Y., Bollivar, D. W. & Bauer, C. E. (1997) Annu. Rev. Genet. 31, 61-89. - PubMed
- Beale, S. I. (1999) Photosynth. Res. 60, 43-73.
- Block, M. A., Tewari, A. K., Albrieux, C., Maréchal, E. & Joyard, J. (2002) Eur. J. Biochem. 269, 240-248. - PubMed
- Porra, R. J., Urzinger, M., Winkler, J., Bubenzer, C. & Scheer, H. (1998) Eur. J. Biochem. 257, 185-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases