Cysteinyl-tRNA(Cys) formation in Methanocaldococcus jannaschii: the mechanism is still unknown - PubMed (original) (raw)
Cysteinyl-tRNA(Cys) formation in Methanocaldococcus jannaschii: the mechanism is still unknown
Benfang Ruan et al. J Bacteriol. 2004 Jan.
Abstract
Most organisms form Cys-tRNA(Cys), an essential component for protein synthesis, through the action of cysteinyl-tRNA synthetase (CysRS). However, the genomes of Methanocaldococcus jannaschii, Methanothermobacter thermautotrophicus, and Methanopyrus kandleri do not contain a recognizable cysS gene encoding CysRS. It was reported that M. jannaschii prolyl-tRNA synthetase (C. Stathopoulos, T. Li, R. Longman, U. C. Vothknecht, H. D. Becker, M. Ibba, and D. Söll, Science 287:479-482, 2000; R. S. Lipman, K. R. Sowers, and Y. M. Hou, Biochemistry 39:7792-7798, 2000) or the M. jannaschii MJ1477 protein (C. Fabrega, M. A. Farrow, B. Mukhopadhyay, V. de Crécy-Lagard, A. R. Ortiz, and P. Schimmel, Nature 411:110-114, 2001) provides the "missing" CysRS activity for in vivo Cys-tRNA(Cys) formation. These conclusions were supported by complementation of temperature-sensitive Escherichia coli cysS(Ts) strain UQ818 with archaeal proS genes (encoding prolyl-tRNA synthetase) or with the Deinococcus radiodurans DR0705 gene, the ortholog of the MJ1477 gene. Here we show that E. coli UQ818 harbors a mutation (V27E) in CysRS; the largest differences compared to the wild-type enzyme are a fourfold increase in the K(m) for cysteine and a ninefold reduction in the k(cat) for ATP. While transformants of E. coli UQ818 with archaeal and bacterial cysS genes grew at a nonpermissive temperature, growth was also supported by elevated intracellular cysteine levels, e.g., by transformation with an E. coli cysE allele (encoding serine acetyltransferase) or by the addition of cysteine to the culture medium. An E. coli cysS deletion strain permitted a stringent complementation test; growth could be supported only by archaeal or bacterial cysS genes and not by archaeal proS genes or the D. radiodurans DR0705 gene. Construction of a D. radiodurans DR0705 deletion strain showed this gene to be dispensable. However, attempts to delete D. radiodurans cysS failed, suggesting that this is an essential Deinococcus gene. These results imply that it is not established that proS or MJ1477 gene products catalyze Cys-tRNA(Cys) synthesis in M. jannaschii. Thus, the mechanism of Cys-tRNA(Cys) formation in M. jannaschii still remains to be discovered.
Figures
FIG. 1.
Scheme of cassette constructs for Deinococcus gene deletions. (A) DR0705 in the D. radiodurans R1 chromosome. (B) DR0705 deletion cassette. (C) DR1670 deletion cassette. Primers indicated above or below the cassette were used for PCR. Sections of the diagrams are labeled as follows: A upstream, 971 bp immediately upstream of the initiation codon of DR0705; A downstream, 806 bp immediately downstream of the termination codon of DR0705; B upstream, 886 bp immediately upstream of the initiation codon of DR1670; B downstream, 957 bp immediately downstream of the termination codon of DR1670.
FIG. 2.
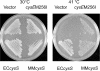
Growth of E. coli cysS(Ts) strain UQ818 at 41°C on a cysteine gradient λ plate. The highest cysteine concentration is in the well (see Materials and Methods).
FIG. 3.
Complementation of E. coli cysS(Ts) strain UQ818 with pCYB1, pCYB-EC_cysS_, pCYB-cysE_M256I, or pCYB-MM_cysS. LB agar containing ampicillin and IPTG was used for growth at 41°C (see Materials and Methods).
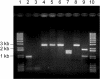
FIG. 4.
Replacement of the D. radiodurans DR0705 gene with the P_katA-npt_ cassette. Lanes 1 and 10, DNA size markers; lanes 2, 4, 6, and 8, DNA from D. radiodurans R1; lanes 3, 5, 7, and 9, DNA from DR0705 deletion strain TNK201. Lanes: 2 and 3, PCR products of primers AFW4 and ARV4; 4 and 5, PCR products of primers AFW2 and ARV3; 6 and 7, _Hin_dIII digestion of DNA in lanes 4 and 5, respectively; 8 and 9, _Xho_I digestion of DNA in lanes 4 and 5, respectively. DR0705 has no restriction sites for _Hin_dIII and _Xho_I, while the npt gene is cut once by both enzymes.
Similar articles
- Methanocaldococcus jannaschii prolyl-tRNA synthetase charges tRNA(Pro) with cysteine.
Ambrogelly A, Ahel I, Polycarpo C, Bunjun-Srihari S, Krett B, Jacquin-Becker C, Ruan B, Köhrer C, Stathopoulos C, RajBhandary UL, Söll D. Ambrogelly A, et al. J Biol Chem. 2002 Sep 20;277(38):34749-54. doi: 10.1074/jbc.M206929200. Epub 2002 Jul 18. J Biol Chem. 2002. PMID: 12130658 - A dual-specificity aminoacyl-tRNA synthetase in the deep-rooted eukaryote Giardia lamblia.
Bunjun S, Stathopoulos C, Graham D, Min B, Kitabatake M, Wang AL, Wang CC, Vivarès CP, Weiss LM, Söll D. Bunjun S, et al. Proc Natl Acad Sci U S A. 2000 Nov 21;97(24):12997-3002. doi: 10.1073/pnas.230444397. Proc Natl Acad Sci U S A. 2000. PMID: 11078517 Free PMC article. - Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis.
Stathopoulos C, Kim W, Li T, Anderson I, Deutsch B, Palioura S, Whitman W, Söll D. Stathopoulos C, et al. Proc Natl Acad Sci U S A. 2001 Dec 4;98(25):14292-7. doi: 10.1073/pnas.201540498. Epub 2001 Nov 20. Proc Natl Acad Sci U S A. 2001. PMID: 11717392 Free PMC article. - Cys-tRNACys formation and cysteine biosynthesis in methanogenic archaea: two faces of the same problem?
Ambrogelly A, Kamtekar S, Sauerwald A, Ruan B, Tumbula-Hansen D, Kennedy D, Ahel I, Söll D. Ambrogelly A, et al. Cell Mol Life Sci. 2004 Oct;61(19-20):2437-45. doi: 10.1007/s00018-004-4194-9. Cell Mol Life Sci. 2004. PMID: 15526152 Review. - Cysteinyl-tRNA formation and prolyl-tRNA synthetase.
Jacquin-Becker C, Ahel I, Ambrogelly A, Ruan B, Söll D, Stathopoulos C. Jacquin-Becker C, et al. FEBS Lett. 2002 Mar 6;514(1):34-6. doi: 10.1016/s0014-5793(02)02331-1. FEBS Lett. 2002. PMID: 11904177 Review.
Cited by
- Evolutionary profiles from the QR factorization of multiple sequence alignments.
Sethi A, O'Donoghue P, Luthey-Schulten Z. Sethi A, et al. Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):4045-50. doi: 10.1073/pnas.0409715102. Epub 2005 Mar 1. Proc Natl Acad Sci U S A. 2005. PMID: 15741270 Free PMC article. - Quality control despite mistranslation caused by an ambiguous genetic code.
Ruan B, Palioura S, Sabina J, Marvin-Guy L, Kochhar S, Larossa RA, Söll D. Ruan B, et al. Proc Natl Acad Sci U S A. 2008 Oct 28;105(43):16502-7. doi: 10.1073/pnas.0809179105. Epub 2008 Oct 22. Proc Natl Acad Sci U S A. 2008. PMID: 18946032 Free PMC article. - Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance.
Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN, Battista JR. Tanaka M, et al. Genetics. 2004 Sep;168(1):21-33. doi: 10.1534/genetics.104.029249. Genetics. 2004. PMID: 15454524 Free PMC article. - The evolutionary history of Cys-tRNACys formation.
O'Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. O'Donoghue P, et al. Proc Natl Acad Sci U S A. 2005 Dec 27;102(52):19003-8. doi: 10.1073/pnas.0509617102. Proc Natl Acad Sci U S A. 2005. PMID: 16380427 Free PMC article. - Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics.
Francklyn CS, Mullen P. Francklyn CS, et al. J Biol Chem. 2019 Apr 5;294(14):5365-5385. doi: 10.1074/jbc.REV118.002956. Epub 2019 Jan 22. J Biol Chem. 2019. PMID: 30670594 Free PMC article. Review.
References
- Ahel, I., C. Stathopoulos, A. Ambrogelly, A. Sauerwald, H. Toogood, T. Hartsch, and D. Söll. 2002. Cysteine activation is an inherent in vitro property of prolyl-tRNA synthetases. J. Biol. Chem. 277:34743-34748. - PubMed
- Ambrogelly, A., I. Ahel, C. Polycarpo, S. Bunjun-Srihari, B. Krett, C. Jacquin-Becker, B. Ruan, C. Köhrer, C. Stathopoulos, U. L. RajBhandary, and D. Söll. 2002. Methanocaldococcus jannaschii prolyl-tRNA synthetase charges tRNAPro with cysteine. J. Biol. Chem. 277:34749-34754. - PubMed
- Battista, J. R. 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51:203-224. - PubMed
- Beuning, P. J., and K. Musier-Forsyth. 2001. Species-specific differences in amino acid editing by class II prolyl-tRNA synthetase. J. Biol. Chem. 276:30779-30785. - PubMed
- Bohman, K., and L. A. Isaksson. 1979. Temperature-sensitive mutants in cysteinyl-tRNA ligase of E. coli K-12. Mol. Gen. Genet. 176:53-55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous