FusionDB: a database for in-depth analysis of prokaryotic gene fusion events - PubMed (original) (raw)
FusionDB: a database for in-depth analysis of prokaryotic gene fusion events
Karsten Suhre et al. Nucleic Acids Res. 2004.
Abstract
FusionDB (http://igs-server.cnrs-mrs.fr/FusionDB/) constitutes a resource dedicated to in-depth analysis of bacterial and archaeal gene fusion events. Such events can provide the 'Rosetta stone' in the search for potential protein-protein interactions, as well as metabolic and regulatory networks. However, the false positive rate of this approach may be quite high, prompting a detailed scrutiny of putative gene fusion events. FusionDB readily provides much of the information required for that task. Moreover, FusionDB extends the notion of gene fusion from that of a single gene to that of a family of genes by assembling pairs of genes from different genomes that belong to the same Cluster of Orthogonal Groups (COG). Multiple sequence alignments and phylogenetic tree reconstruction for the N- and C-terminal parts of these 'COG fusion' events are provided to distinguish single and multiple fusion events from cases of gene fission, pseudogenes and other false positives. Finally, gene fusion events with matches to known structures of heterodimers in the Protein Data Bank (PDB) are identified and may be visualized. FusionDB is fully searchable with access to sequence and alignment data at all levels. A number of different scores are provided to easily differentiate 'real' from 'questionable' cases, especially when larger database searches are performed. FusionDB is cross-linked with the 'Phylogenomic Display of Bacterial Genes' (PhydBac) online web server. Together, these servers provide the complete set of information required for in-depth analysis of non-homology-based gene function attribution.
Figures
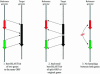
Figure 1
Criteria for a putative gene fusion event based on a mutual best match criteria (see text for details).
Figure 2
Screenshot of FusionDB full-page output for a query to COG2080 and examples of some related information that can be obtained through this page. PhydBac (
http://igs-server.cnrs-mrs.fr/phydbac/
) is the ‘Phylogenomic Display of Bacterial Genes’ online web tool. In the top of the ‘COG fusion alignment’, N- and C- terminal genes are presented in red and green, respectively, fusion ORFs are in black. The alignment of the merged genes with the fusion genes is presented below. A colour scale ranging from green over yellow to red represents the EMBOSS plotcon score for this ‘merged alignment’. The ‘phylogenetic trees’ are based on the N- and the C-terminal ‘COG fusion alignments’, respectively. Genomes in which fusion events occurred are highlighted in red in the trees. The ‘alignment to the PDB’ is a representation of the T-Coffee alignment core index of the reference genes (top row), the fusion ORF (middle row) and the sequence of the heterodimer (bottom row), warmer colours indicating a higher confidence in the alignment quality. PDBsum (
http://www.biochem.ucl.ac.uk/bsm/pdbsum/
) is a database of the known 3D structures of proteins and nucleic acids.
Similar articles
- Inference of gene function based on gene fusion events: the rosetta-stone method.
Suhre K. Suhre K. Methods Mol Biol. 2007;396:31-41. doi: 10.1007/978-1-59745-515-2_3. Methods Mol Biol. 2007. PMID: 18025684 - Phydbac2: improved inference of gene function using interactive phylogenomic profiling and chromosomal location analysis.
Enault F, Suhre K, Poirot O, Abergel C, Claverie JM. Enault F, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W336-9. doi: 10.1093/nar/gkh365. Nucleic Acids Res. 2004. PMID: 15215406 Free PMC article. - A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm.
Podell S, Gaasterland T, Allen EE. Podell S, et al. BMC Bioinformatics. 2008 Oct 7;9:419. doi: 10.1186/1471-2105-9-419. BMC Bioinformatics. 2008. PMID: 18840280 Free PMC article. - Comparative Genomics for Prokaryotes.
Setubal JC, Almeida NF, Wattam AR. Setubal JC, et al. Methods Mol Biol. 2018;1704:55-78. doi: 10.1007/978-1-4939-7463-4_3. Methods Mol Biol. 2018. PMID: 29277863 Review. - What we have learned about prokaryotes from structural genomics.
Frishman D. Frishman D. OMICS. 2003 Summer;7(2):211-24. doi: 10.1089/153623103322246601. OMICS. 2003. PMID: 14506850 Review.
Cited by
- Identification of genomic features using microsyntenies of domains: domain teams.
Pasek S, Bergeron A, Risler JL, Louis A, Ollivier E, Raffinot M. Pasek S, et al. Genome Res. 2005 Jun;15(6):867-74. doi: 10.1101/gr.3638405. Epub 2005 May 17. Genome Res. 2005. PMID: 15899966 Free PMC article. - Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information.
Ovchinnikov S, Kamisetty H, Baker D. Ovchinnikov S, et al. Elife. 2014 May 1;3:e02030. doi: 10.7554/eLife.02030. Elife. 2014. PMID: 24842992 Free PMC article. - 'Conserved hypothetical' proteins: prioritization of targets for experimental study.
Galperin MY, Koonin EV. Galperin MY, et al. Nucleic Acids Res. 2004 Oct 12;32(18):5452-63. doi: 10.1093/nar/gkh885. Print 2004. Nucleic Acids Res. 2004. PMID: 15479782 Free PMC article. Review. - Structure of the bifunctional methyltransferase YcbY (RlmKL) that adds the m7G2069 and m2G2445 modifications in Escherichia coli 23S rRNA.
Wang KT, Desmolaize B, Nan J, Zhang XW, Li LF, Douthwaite S, Su XD. Wang KT, et al. Nucleic Acids Res. 2012 Jun;40(11):5138-48. doi: 10.1093/nar/gks160. Epub 2012 Feb 22. Nucleic Acids Res. 2012. PMID: 22362734 Free PMC article. - ProtFus: A Comprehensive Method Characterizing Protein-Protein Interactions of Fusion Proteins.
Tagore S, Gorohovski A, Jensen LJ, Frenkel-Morgenstern M. Tagore S, et al. PLoS Comput Biol. 2019 Aug 22;15(8):e1007239. doi: 10.1371/journal.pcbi.1007239. eCollection 2019 Aug. PLoS Comput Biol. 2019. PMID: 31437145 Free PMC article.
References
- Galperin M.Y. and Koonin,E.V. (2000) Who’s your neighbor? New computational approaches for functional genomics. Nat. Biotechnol., 18, 609–613. - PubMed
- Sali A. (1999) Functional links between proteins. Nature, 402, 23–26. - PubMed
- Marcotte E.M. (2000) Computational genetics: finding protein function by nonhomology methods. Curr. Opin. Struct. Biol., 10, 359–365. - PubMed
- Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. - PubMed