Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium - PubMed (original) (raw)
Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium
Christopher S Carlson et al. Am J Hum Genet. 2004 Jan.
Abstract
Common genetic polymorphisms may explain a portion of the heritable risk for common diseases. Within candidate genes, the number of common polymorphisms is finite, but direct assay of all existing common polymorphism is inefficient, because genotypes at many of these sites are strongly correlated. Thus, it is not necessary to assay all common variants if the patterns of allelic association between common variants can be described. We have developed an algorithm to select the maximally informative set of common single-nucleotide polymorphisms (tagSNPs) to assay in candidate-gene association studies, such that all known common polymorphisms either are directly assayed or exceed a threshold level of association with a tagSNP. The algorithm is based on the r(2) linkage disequilibrium (LD) statistic, because r(2) is directly related to statistical power to detect disease associations with unassayed sites. We show that, at a relatively stringent r(2) threshold (r2>0.8), the LD-selected tagSNPs resolve >80% of all haplotypes across a set of 100 candidate genes, regardless of recombination, and tag specific haplotypes and clades of related haplotypes in nonrecombinant regions. Thus, if the patterns of common variation are described for a candidate gene, analysis of the tagSNP set can comprehensively interrogate for main effects from common functional variation. We demonstrate that, although common variation tends to be shared between populations, tagSNPs should be selected separately for populations with different ancestries.
Figures
Figure 1
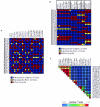
Common variation and LD in European Americans at BDKRB2. At the BDKRB2 gene, 22 SNPs with
_MAF_>10%
were described for the European American samples (A). Patterns of genotype at each SNP are shown as a visual genotype plot, in which each column represents a site and each row represents a sample. Genotype is color coded, as shown, with SNPs presented in the order they were identified across the gene. Patterns of genotype are clearly similar for many SNPs (e.g., sites 10922 and 12574) but not necessarily for adjacent SNPs. The same data are shown in panel B, with the order of SNPs rearranged such that each SNP is adjacent to SNPs with similar patterns of genotype. Among the 22 SNPs, the LD-based SNP-selection algorithm identified five bins of tagSNPs at an _r_2 threshold of 0.5. tagSNP bins are boxed (B). The LD statistic _r_2 describes the similarity of pattern between pairs of polymorphic sites: pairwise _r_2 between SNPs is shown for the same order of SNPs as in panel B, and bins of SNPs with similar patterns are visible as reddish triangles above the diagonal (C).
Figure 2
tagSNPs per gene, with threshold
_r_2>0.5
and
_MAF_>10%
. The complete genomic region of 100 genes was resequenced in 24 unrelated African American and 23 unrelated European American samples. Within each population, tagSNPs were selected from all SNPs with
_MAF_>10%
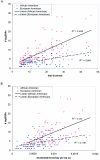
at an _r_2 threshold of 0.5. A, The number of tagSNPs selected in each gene under these parameters, plotted against the size of the genomic region for each gene. Although there is a clear trend toward more tagSNPs in larger genes, there is considerable variance in the required tagSNP density in both populations. B, The number of tagSNPs selected in each gene, plotted against nucleotide diversity (π) per base pair. Thus, variance in tagSNP density between genes reflects both variation in nucleotide diversity and variation in the average extent of LD within genes. Within each gene, a greater number of tagSNPs is generally required in the African American population, reflecting both greater nucleotide diversity and shorter range LD, relative to the European American population.
Figure 3
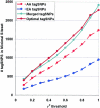
Total tagSNP bins in 100 genes, versus threshold _r_2. At each _r_2 threshold, tagSNP bins were identified for 100 genes within African American (“AA tagSNPs”) and European American (“EA tagSNPs”) populations. As expected, more tagSNP bins were identified in African American samples than in European American samples. To measure the effects of population stratification on the LD-select algorithm, tagSNPs were also selected from merged African American and European American populations (“Merged tagSNPs”). The minimal set of tagSNPs relevant to both populations was also assembled at each _r_2 threshold as the union of the tagSNP sets selected in each subpopulation (“Optimal tagSNPs”); this set was larger than the tagSNP set in either subpopulation alone but considerably smaller than the sum of the population-specific site sets, reflecting the fact that many (but not all) tagSNPs were useful in both populations.
Figure 4
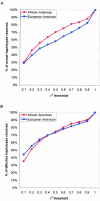
The relationship between LD-selected tagSNPs and haplotypes. For each gene, haplotypes were inferred computationally. Results are shown as the fraction of haplotypes resolved using only LD-selected tagSNPs, relative to haplotypes resolved using all common SNPs. Results are shown across a range of _r_2 values in each population (A). The effective number of haplotypes weights the number of haplotypes by frequency, with common haplotypes more heavily weighted. For each gene, the fraction of effective haplotypes resolved using only LD-selected tagSNPs, relative to effective haplotypes resolved, by use of all common SNPs is shown across a range of _r_2 values in each population (B). For _r_2 thresholds >0.5, >80% of effective haplotypes were resolved, demonstrating how, at adequately stringent _r_2 thresholds, LD-selected tagSNPs efficiently resolve common haplotypes.
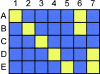
Figure 5
tagSNP bins and the evolutionary relationships between haplotypes. A hypothetical nonrecombinant region with five existing haplotypes is shown, with each row (A–E) representing a haplotype and each column (1–7) representing an SNP with a unique pattern of alleles. The common allele is shown as blue and the rare allele as yellow. There are five possible patterns (1–5) that are haplotype specific, and two (6 and 7) that are specific to clades of related haplotypes. LD-based tagSNP selection at an adequately stringent _r_2 threshold would identify all seven patterns in this hypothetical region. Thus, directly testing LD-selected tagSNPs can identify disease associations with either specific haplotypes or with clades of related haplotypes.
Similar articles
- Efficient selection of tagging single-nucleotide polymorphisms in multiple populations.
Howie BN, Carlson CS, Rieder MJ, Nickerson DA. Howie BN, et al. Hum Genet. 2006 Aug;120(1):58-68. doi: 10.1007/s00439-006-0182-5. Epub 2006 May 6. Hum Genet. 2006. PMID: 16680432 - Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates.
Pimenoff VN, Laval G, Comas D, Palo JU, Gut I, Cann H, Excoffier L, Sajantila A. Pimenoff VN, et al. Pharmacogenet Genomics. 2012 Dec;22(12):846-57. doi: 10.1097/FPC.0b013e32835a3a6d. Pharmacogenet Genomics. 2012. PMID: 23089684 - A comprehensive analysis of common genetic variation in prolactin (PRL) and PRL receptor (PRLR) genes in relation to plasma prolactin levels and breast cancer risk: the multiethnic cohort.
Lee SA, Haiman CA, Burtt NP, Pooler LC, Cheng I, Kolonel LN, Pike MC, Altshuler D, Hirschhorn JN, Henderson BE, Stram DO. Lee SA, et al. BMC Med Genet. 2007 Dec 1;8:72. doi: 10.1186/1471-2350-8-72. BMC Med Genet. 2007. PMID: 18053149 Free PMC article. - On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.
Garner C, Slatkin M. Garner C, et al. Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review. - [Linkage disequilibrium in the human genome and its exploitation].
Kharrat N, Rebaï M, Rebaï A. Kharrat N, et al. Arch Inst Pasteur Tunis. 2005;82(1-4):9-21. Arch Inst Pasteur Tunis. 2005. PMID: 16929750 Review. French.
Cited by
- Transferability and fine mapping of genome-wide associated loci for lipids in African Americans.
Adeyemo A, Bentley AR, Meilleur KG, Doumatey AP, Chen G, Zhou J, Shriner D, Huang H, Herbert A, Gerry NP, Christman MF, Rotimi CN. Adeyemo A, et al. BMC Med Genet. 2012 Sep 21;13:88. doi: 10.1186/1471-2350-13-88. BMC Med Genet. 2012. PMID: 22994408 Free PMC article. - No association between germline variation in catechol-O-methyltransferase and colorectal cancer survival in postmenopausal women.
Passarelli MN, Newcomb PA, Makar KW, Burnett-Hartman AN, Phipps AI, David SP, Hsu L, Harrison TA, Hutter CM, Duggan DJ, White E, Chan AT, Peters U. Passarelli MN, et al. Menopause. 2014 Apr;21(4):415-20. doi: 10.1097/GME.0b013e31829e498d. Menopause. 2014. PMID: 23880798 Free PMC article. - A non-synonymous coding variant (L616F) in the TLR5 gene is potentially associated with Crohn's disease and influences responses to bacterial flagellin.
Sheridan J, Mack DR, Amre DK, Israel DM, Cherkasov A, Li H, Grimard G, Steiner TS. Sheridan J, et al. PLoS One. 2013 Apr 11;8(4):e61326. doi: 10.1371/journal.pone.0061326. Print 2013. PLoS One. 2013. PMID: 23593463 Free PMC article. - Assessing effectiveness of many-objective evolutionary algorithms for selection of tag SNPs.
Moqa R, Younas I, Bashir M. Moqa R, et al. PLoS One. 2022 Dec 8;17(12):e0278560. doi: 10.1371/journal.pone.0278560. eCollection 2022. PLoS One. 2022. PMID: 36480538 Free PMC article. - Identification of haplotype tag single nucleotide polymorphisms within the receptor for advanced glycation end products gene and their clinical relevance in patients with major trauma.
Zeng L, Zhang AQ, Gu W, Zhou J, Zhang LY, Du DY, Zhang M, Wang HY, Yan J, Yang C, Jiang JX. Zeng L, et al. Crit Care. 2012 Jul 24;16(4):R131. doi: 10.1186/cc11436. Crit Care. 2012. PMID: 22827914 Free PMC article.
References
Electronic-Database Information
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (Accession numbers for all genes are listed in .)
- HaploBlockFinder, http://cgi.uc.edu/cgi-bin/kzhang/haploBlockFinder.cgi/
- Pharmacogenetics and Risk of Cardiovascular Disease Project, http://droog.gs.washington.edu/parc/
- Phred/Phrap/Consed System Web Site, http://www.phrap.org/
References
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalayanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–23810.1038/10290 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HL066642/HL/NHLBI NIH HHS/United States
- N01ES15478/ES/NIEHS NIH HHS/United States
- R37 MH059520/MH/NIMH NIH HHS/United States
- MH59520/MH/NIMH NIH HHS/United States
- HL66682/HL/NHLBI NIH HHS/United States
- HL66642/HL/NHLBI NIH HHS/United States
- R01 MH059520/MH/NIMH NIH HHS/United States
- U01 HL066682/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials